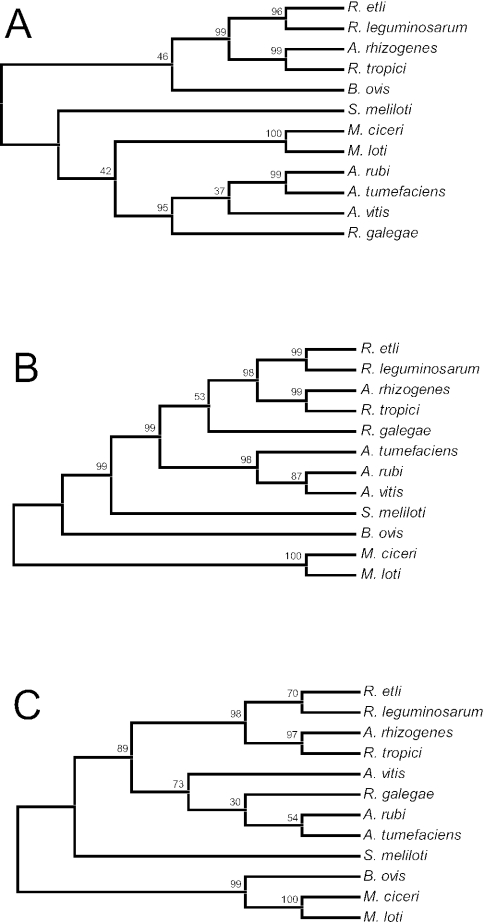
FIG. 2.
Unrooted maximum-parsimony trees based on single-gene sequences from 12 species representing five genera (Agrobacterium, Brucella, Mesorhizobium, Rhizobium, and Sinorhizobium) within the order Rhizobiales. Trees were generated by a heuristic mini-min tree search option with a search factor of 2. The percentage bootstrap support at each internal node is based on 1,000 replicate trees. (A) 16S rRNA nucleotide sequence tree. The total alignment length for the analysis was 1,401 bp. (B) dnaK nucleotide sequence tree. The total alignment length for the analysis was 1,917 bp. (C) Inferred DnaK amino acid sequence tree. The total alignment included 642 amino acid residues.