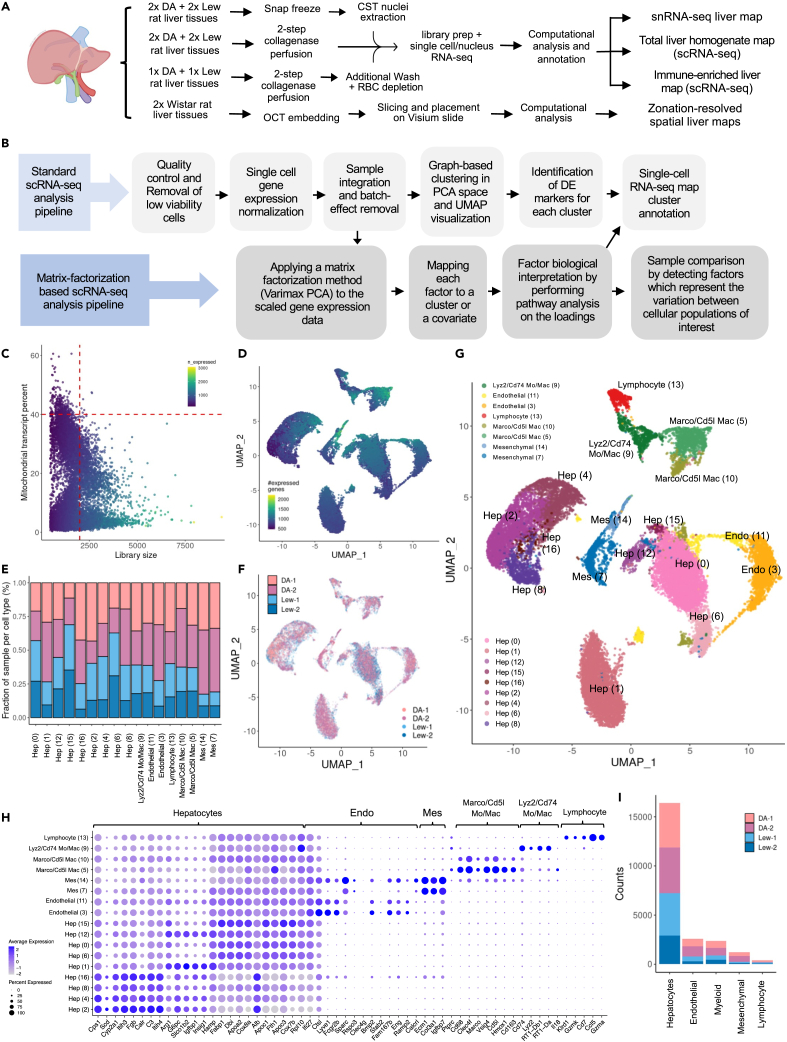
Figure 1.
scRNA-seq profiling of rat liver reveals 17 distinct cell populations
(A) Overview of single-cell RNA-seq pipeline, including both the experimental and analysis workflows.
(B) Major steps of the standard and matrix factorization-based single-cell RNA-seq data analysis pipeline.
(C) Viable cell selection for a Lewis rat liver sample (LEW-1) based on library size and mitochondrial transcript proportion shown as an example. High-quality cells were identified from the single-cell libraries having a minimum library size of 1500 transcripts and a maximum of 40% mitochondrial transcript proportion.
(D) UMAP (uniform manifold approximation and projection for dimension reduction) plot of four rat samples including 2 samples from each Dark agouti (DA) and Lewis (LEW) rat strains. Cells are colored by the number of expressed genes, with lighter colors indicating higher gene counts.
(E) Bar plot indicating the relative contribution of input samples to each cluster. All samples are represented in each cluster.
(F) UMAP projection of cells labeled based on the input sample indicates that cells from different samples have been well-integrated and clusters represent cell-type differences rather than sample-specific variations.
(G) UMAP projection of four total liver homogenate rat samples (each point represents a single cell) where cells that share similar transcriptome profiles are grouped by colors representing unsupervised clustering results. The legend indicates the unique color representing the cell-type annotation of each cluster. The cluster number is shown within the curved brackets.
(H) Dot plot indicating the relative expression of marker genes in each population. The size of the circle indicates the percentage of cells in each population which express the marker of interest, and the color represents the average expression value of the marker.
(I) The number of cells in each major cell type population colored by the contribution of each input sample. RBC: red blood cell, PCA: principal-component analysis, DE: differentially expressed, QC: quality control, Mac: macrophage, Mo: monocyte, Endo: endothelial, Mes: mesenchymal, Hep: hepatocyte.