Figure 2.
SnRNA-seq and spatial transcriptomic profiling of the rat liver resolves hepatocyte zonation
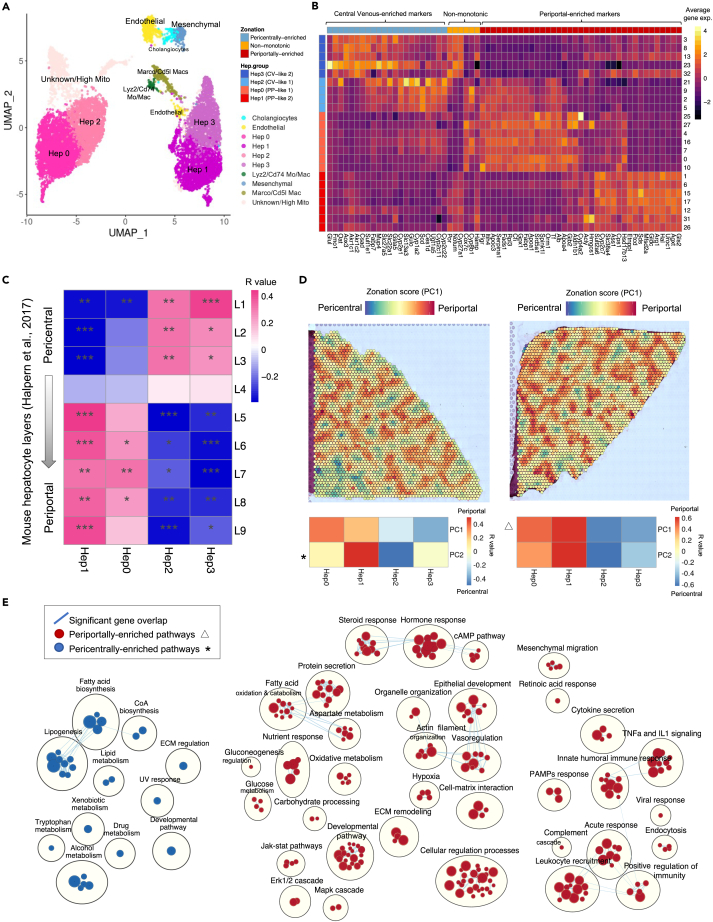
Four additional rat liver samples were added and sequenced using snRNA-seq to better characterize hepatocyte and cholangiocyte populations and verify the strain variations identified based on the scRNA-seq TLH map. (A) UMAP projection of four snRNA-seq samples where cells are colored based on cell-type annotation.
(B) Heatmap representing the average gene expression of zonated genes based on spatial data within the snRNA-seq clusters.
(C) Pearson correlation between the average gene expression of the genes across snRNA-seq hepatocyte clusters and the nine layers of mouse liver cells was calculated (see STAR Methods). Mouse liver layer-9 is more periportal and layer 1 is pericentral. Red represents a positive correlation, and blue represents a negative correlation. (∗: p value <0.05, ∗∗: p value <0.01, ∗∗∗: p value <0.001).
(D) Projection of zonation signature scores, captured by PC1, across the spatial transcriptomics spots of two healthy Wistar rat liver cryosections. The top negatively loaded genes in PC2 (and PC1) of both samples are enriched in pericentral markers, and the top positively loaded genes in PC1 (and PC2) factors are enriched in periportal markers. Red and blue represent high periportal and pericentral zonation scores, respectively. The two heatmaps represent the Pearson correlation between the zonation factors PC1 and PC2 and the average expression of snRNA-seq hepatocyte clusters. Both PC1 and PC2 are positively correlated with PP-like clusters Hep0 and Hep1 and negatively correlated with CV-like clusters Hep2 and Hep3. The asterisk and triangle symbols indicate the factors used for pathway enrichment analysis.
(E) Pathway enrichment analysis using GSEA (gene set enrichment analysis) to examine active cellular pathways in periportal and central venous regions of the healthy rat liver based on spatial PC1 and PC2 loadings visualized as an enrichment map. The pathways enriched in the pericentral and periportal areas are based on PC2 (asterisk) of liver cryosections-A (left) and PC1 (triangle) of liver cryosections-B (right) respectively. Each circle represents a gene ontology (GO) biological process term. The size of the circles represents the number of genes in that pathway and blue lines indicate significant gene overlap.