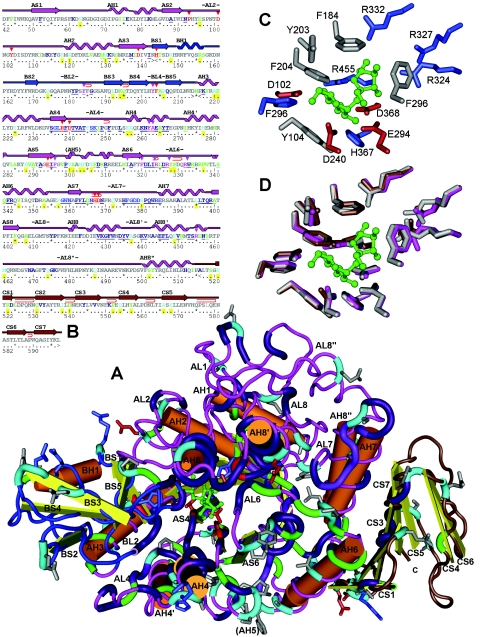
FIG.2.
Predicted secondary and 3D structure of the P. dispersa UQ68J SI. The colors of the peptide backbone indicate domains (pink, A [TIM barrel] domain; blue, B subdomain; brown, C domain). (A) In the 3D structure, the core of the TIM barrel is under the loop marked AL6, and the eight beta sheets point upward. The substrate sucrose molecule (green) is located near the top of the barrel and is surrounded by conserved glycolytic residues (red). Other expanded sections of the peptide backbone represent residues that are conserved among sucrose isomerases (purple), unique to the Pseudomonas trehalulose synthase (green), or unique to the UQ68J isomaltulose synthase among residues functionally conserved across other sucrose isomerases (light blue). Amino acid side chains are shown only for the conserved active site and unique UQ68J residues (different colors indicate different charges). Prominent beta sheets (S), alpha helices (H), and connecting loops (L) are numbered by domain from the N terminus. (B) For the secondary structure the same numbering and color coding are used. Residues unique to UQ68J are indicated by yellow, conserved strings near the active site are underlined, residues closest to bound glucose and fructose moieties are indicated by red and pink arrows, glycolytic core residues are underlined with wavy lines, and tight folds in loop regions are indicated by hairpin symbols. (C) Amino acids around the UQ68J active site. (D) Overlaid active sites for SIs from strains UQ68J (pink), LX3 (brown), and WAC2928 (grey).