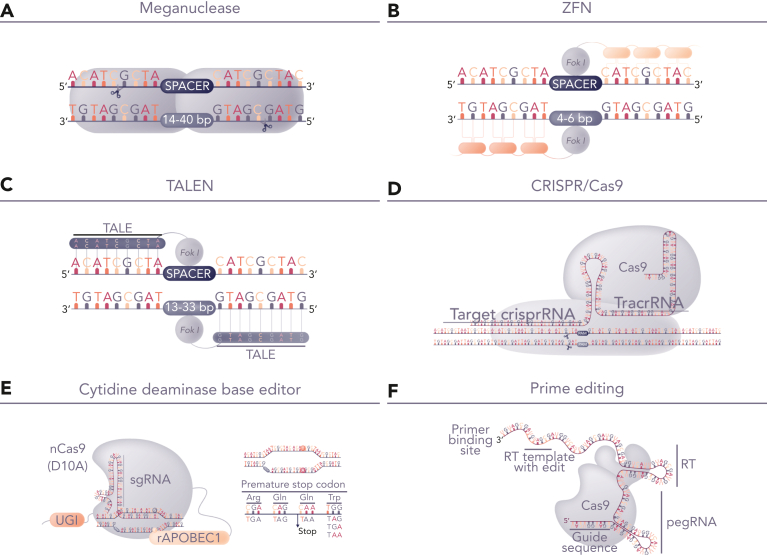
Figure 1.
Genome-editing platforms for T-cell modification. (A-D) Editing tools for highly specific dsDNA cleavage comprise either protein-based DNA recognition molecules including (A) homing endonucleases (meganuclease), (B) zinc finger nucleases, (C) TALENs, (D) RNA-guided nucleases exemplified by CRISPR/Cas9. After DNA cleavage, repair by nonhomologous end joining offers the prospect of gene-knockout with indel signatures and alternative homologous repair based repair, which can be exploited for site-specific transgene insertion. The latter requires delivery of template DNA flanked by homology arms and allows placement of CAR genes under the transcriptional control of endogenous transcriptional machinery. Alternatively, (E) base editors use nickase-restricted Cas9 variants, fused to cytidine deaminase or adenosine deaminase for highly targeted C>T or A>G base conversion. Cytidine base editor (CBE) has been used for multiplexed gene knockout, avoiding translocations usually encountered following nuclease activity. (F) Emerging prime editing offers the prospect of gene editing through localized template repair by fusing reverse transcriptase to deactivated Cas9. dsDNA, double-stranded DNA.