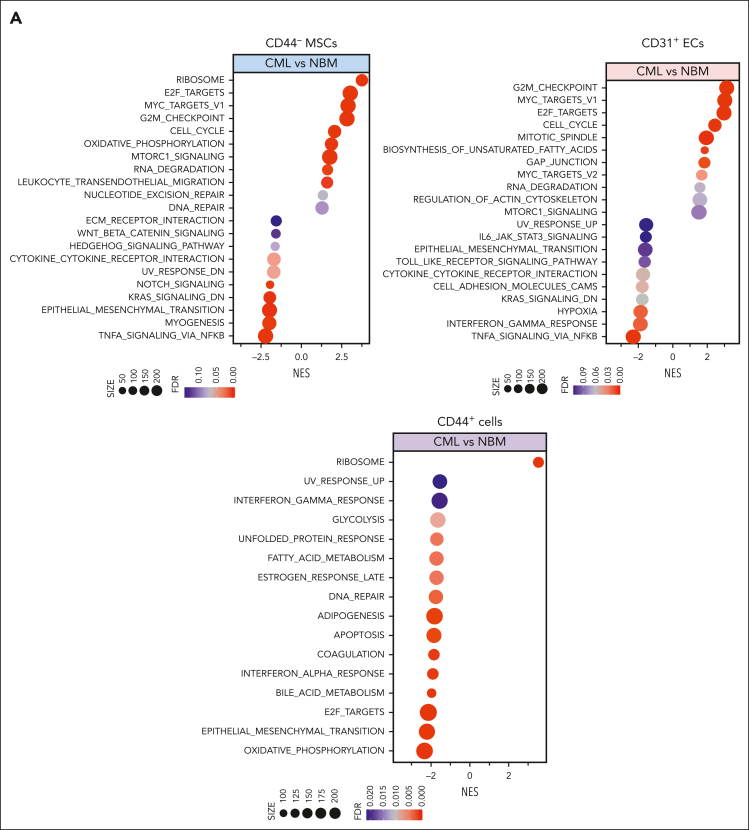
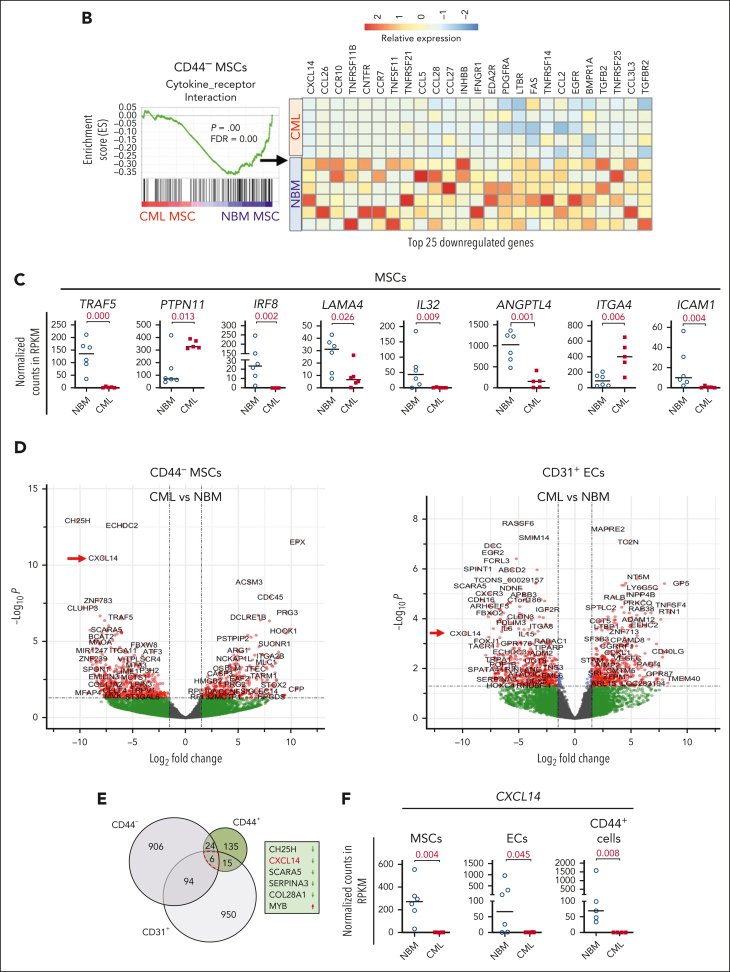
Figure 2.
RNA sequencing reveals molecular profiles of CML BM cellular niches. RNA sequencing was performed on FACS-sorted BM CD44– MSCs, CD31+ ECs, and CD44+ mature stromal cells from 5 patients with CML and 6 healthy BM donors. The false discovery rate (FDR) q value (FDR-q value) represents the false discovery rate of the P values. Gene set enrichment analysis (GSEA) was carried out on the RNA sequencing data to identify differentially expressed genes in the CML-derived stromal cells. (A) GSEA reveals differentially expressed gene sets in the CML CD44– MSCs, CD31+ ECs, and CD44+ mature stromal cells. (B) GSEA plot showing the downregulation of genes involved in the cytokine-receptor interaction in CML MSCs and the heatmap of the top 25 altered genes in this pathway. (C) Selected inflammatory cytokine and adhesion molecule expression in the CML MSCs. (D) Volcano plots showing the altered genes in the CML BM MSCs and ECs in comparison with their healthy counterparts. The red arrows highlight CXCL14 expression. (E) Venn diagram summarizing the commonly differentially expressed genes within the CML CD44– MSCs, CD31+ ECs, and CD44+ mature stromal cells. The numbers in the panel represent the genes that are significantly (P < .01 and FDR < 0.10) altered. (F) Downregulation of CXCL14 transcripts in the MSCs, CD44+ mature stromal cells, and ECs from patients with CML. Horizontal lines represent median values. The data were from 5 patients with CML and 6 normal BM donors. See also supplemental Figures 3-5. RPKM, reads per kilobase of transcript, per million mapped reads.