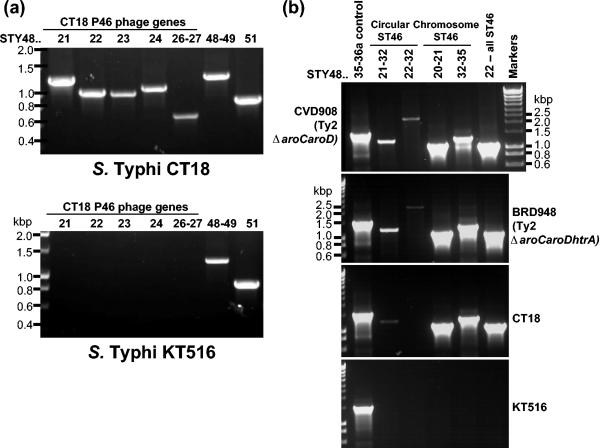
FIG. 3.
(a) PCR analysis confirms the absence of the CT18 P4 phage in Salmonella serovar Typhi KT516. The primers used for these analyses are as follows (see also Table S1 in the supplemental material): lane 1, primer pair 1-3; lane 2, primer pair 4-5; lane 3, primer pair 6-7; lane 4, primer pair 8-9; lane 5, primer pair 10-11; lane 6, primer pair 12-13; lane 7, primer pair 14-15. The CT18 genome numbers for each gene (or genes) amplified and the five PCR products that lie within the CT18 P4 phage (STY4821-24 and STY4826-27) are noted above the gel. PCR analysis of Salmonella serovar Typhi KT516 confirms the observation, made using microarrays (Fig. 2), that this strain does not contain an intact CT18-like P4 phage, whereas transposase STY4848 and the helicase-containing region (STY4849 and STY4851) are present. (b) Circularized ST46 phage DNA was detected by PCR with genomic DNA from Salmonella serovar Typhi strains BRD948, CVD908, or CT18 but not with KT516, which is missing ST46. The CT18 genome numbers for each gene (or genes) amplified are noted above the gel. Whether the primers were designed to amplify chromosomal and/or circular phage DNA products is indicated. The primers used are detailed in Materials and Methods.