Abstract
The zinc metalloprotease EmpA is a virulence factor in the fish pathogen Vibrio anguillarum. Previous studies have shown that two strains of V. anguillarum regulate empA differently. Strain M93Sm exhibits protease activity only in the presence of fish gastrointestinal mucus, while protease activity is detected in NB10 culture supernatant under all stationary-phase conditions. In this study, we use real-time reverse transcription-PCR to show that even in conditions where no protease activity is detected, empA transcription occurs. Western blot analysis revealed that EmpA is secreted as a ∼48-kDa proenzyme and that activation occurs extracellularly by the removal of a ∼10-kDa peptide. The presence of stable extracellular pro-EmpA in M93Sm culture supernatants suggests that activation of EmpA is not autolytic.
The marine bacterium Vibrio anguillarum is the causative agent of vibriosis, a systemic disease of fish characterized by hemorrhagic septicemia (1). Outbreaks of vibriosis result in high mortality rates of infected fish, and it is a major obstacle to the spread of commercial aquaculture (1).
The zinc metalloprotease EmpA has been identified as a virulence factor for V. anguillarum (2, 7, 8). EmpA is a structural homologue to the secreted LasB elastase of Pseudomonas aeruginosa (7). The gastrointestinal (GI) tract has been shown to be a route of entry for V. anguillarum into fish hosts (9, 10). Incubation of V. anguillarum in gastrointestinal mucus specifically induces a number of proteins, including EmpA (3, 4). Denkin and Nelson (2) showed that the induction of protease activity by GI mucus in stationary-phase cultures varied between two strains of V. anguillarum. Strain M93Sm demonstrated protease activity only in the presence of mucus, while strain NB10 expressed protease activity under all stationary-phase conditions with and without mucus.
In this report, we further describe the differences between NB10 and M93Sm in the regulation of EmpA and show that EmpA is under both transcriptional and posttranslational control. Differences in the induction of empA and the role of gastrointestinal mucus were examined using real-time reverse transcription-PCR (RT-PCR). Western blot analysis was used to describe the secretion and processing of EmpA.
MATERIALS AND METHODS
Strains and culture conditions.
Two wild-type strains of V. anguillarum, NB10 (7) and M93Sm (3), were used in this study. Strain M03, a streptomycin- and chloramphenicol-resistant (Smr Cmr) rpoS mutant derived from M93Sm (2) was also used. Cultures were routinely grown in Luria-Bertani broth plus 2% NaCl (LB20) (4) with the addition of the appropriate antibiotics on a rotary shaker at 27°C. Experimental media included LB20 and nine-salt solution (NSS) plus 200 μg of salmon GI mucus protein ml−1 (NSSM) (4). For experiments, 16-h cultures of V. anguillarum (grown at 27°C) were centrifuged (9,000 × g, 5 min, 4°C), washed twice in NSS, and resuspended at the appropriate cell densities in either LB20 or NSSM. Cell densities were determined by serial dilution and plating. Streptomycin was used at a concentration of 200 μg ml−1, and chloramphenicol was used at 5 μg ml−1.
Measurement of protease activity.
Protease activity was quantified using the azocasein method of Windle and Kelleher (12), as modified by Denkin and Nelson (3).
RNA extractions.
Total RNA was isolated using the RNeasy purification kit (QIAGEN, Valencia, Calif.) and treated with RQ1 RNase-free DNase and RNasin according to the manufacturer's specifications (Promega, Madison, Wis.).
Real-time RT-PCR.
Real-time quantitative RT-PCR (qRT-PCR) was performed using an Mx4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, Calif.). Primers were SYBR2empA-F (5′-TCA AAT CTT CCG GTG GCT TAC GTT-3′) and SYBR2empA-R (5′-GCG CGA TTA AAC ACA CCA CTT GAA-3′). These amplify a 124-bp fragment of the empA gene of V. anguillarum. Quantitation of empA mRNA copy number was performed using Brilliant SYBR Green Single-Step QRT-PCR Master Mix (Stratagene) with 10 ng of total RNA in 50-μl reaction mixtures. The thermal profile was 50°C for 30 min, 95°C for 15 min, and then 40 cycles of 95°C for 30 s and 55°C for 30 s. Fluorescence was measured at the end of the 55°C-step every cycle. Samples were run in triplicate plus one no-RT control.
Preparation of protein extracts.
Supernatant and washed whole-cell proteins were collected from V. anguillarum cultures grown for 16 h in either LB20 or NSSM. Aliquots (1 ml) of the cultures were centrifuged (9,000 × g, 10 min, 4°C) to pellet cells, and supernatants were filtered through 0.22-μm-pore-size MillexGP cartridge filters (Millipore, Billerica, Mass.). Protein was precipitated with 15% tricholoroacetic acid (0°C for 1 h), pelleted by centrifugation (12,000 × g, 15 min, 4°C), rinsed twice with acetone, and resuspended in 15 μl of 1× phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, and 1.5 mM KH2PO4). Cellular protein was collected by washing cell pellets twice with NSS, resuspending them in 0.5 ml of 1× PBS, and disrupting the cells by sonication for 1 min with a sonic dismembrator (Fisher Scientific, Swanee, Ga.). Supernatant proteins and sonicated cells were stored at −70°C until use.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis.
Washed whole-cell and supernatant protein samples were separated in 12% Tris-HCl precast-ready gels (Bio-Rad, Hercules, Calif.). Prestained low-range-molecular-mass standards (Bio-Rad) and 2× Laemmli sample buffer (Sigma, St. Louis, Mo.) were used in all gels. Protein loaded in each lane was either 0.7 ml of the original LB20 or 1.5 ml of the NSSM cultures. Gels were either stained in Bio-Safe Coomassie (Bio-Rad) or transferred to nitrocellulose membranes for immunoblotting using the mini-Protean II system (Bio-Rad). Transfers were performed as described by Towbin et al. (11) at 100 V for 1.5 h. Nitrocellulose membranes were blocked, as described by Girouard et al. (5), with the addition of 5% skim milk. Detection of EmpA bands was done with rabbit anti-P. aeruginosa LasB elastase antibodies at a dilution of 1:7,000. Rabbit anti-LasB antibody was detected with horseradish peroxidase-conjugated goat anti-rabbit antibody (Chemicon, Temecula, Calif.) at a dilution of 1:5,000 and was visualized with a DAB tetrahydrochloride kit (ICN Biochemicals, Aurora, Ohio).
RESULTS AND DISCUSSION
Denkin and Nelson (3) showed that V. anguillarum strain NB10 exhibits EmpA protease activity under all stationary-phase conditions in both LB20 and NSSM, while M93Sm exhibits protease activity only when grown in NSSM, not in LB20. The interpretation of this and other supporting data was that the salmon GI mucus in NSSM acts to induce protease activity in M93Sm, not to activate preexisting but inactive protease (3). The role of mucus as an inducer was further supported by Northern blot analysis showing that M93Sm did not appear to transcribe empA mRNA when grown in LB20 (2).
To further characterize differences in empA transcription, we examined the levels of empA mRNA in NB10 and M93Sm using qRT-PCR. Cultures of NB10 and M93Sm grown for 16 h in LB20 were resuspended at ∼2 × 109 CFU ml−1 in fresh LB20 or NSSM and incubated at 27°C, and samples for RNA isolation were taken at 180 min. Both strains produced empA mRNA when cultured in either LB20 or NSSM (Table 1). For each strain, comparisons of culture conditions showed that NSSM-grown NB10 cells expressed about five times more empA message than LB20-grown NB10 cells, while NSSM-grown M93Sm cells produced about two times more empA message than LB20-grown M93Sm cells (Table 1). Comparisons between the two strains for identical culture conditions showed that LB20-grown NB10 cells produced threefold more empA transcript than did LB20-grown M93Sm cells, while NSSM-grown NB10 cells has 7.5-fold more empA message than NSSM-grown M93Sm cells. The lowest transcript levels were found in LB20-grown M93Sm cells, for which it was over 150 times greater than values found in M03, an rpoS-null mutant known not to express EmpA (2). These results confirm that NB10 and M93Sm regulate empA differentially. However, the presence of empA message in LB20-grown M93Sm cells reveals that the difference in expression between the strains is not solely due to differential induction of empA transcription.
TABLE 1.
qRT-PCR quantification of empA mRNA transcript from M93Sm and NB10 cells incubated in LB20 and NSSM for 180 mina
| Strain | No. of copies of empA transcripts
|
Relative increase from LB20 to NSSM | |
|---|---|---|---|
| LB20 | NSSM | ||
| M93Sm | (3.6 ± 1.0) × 105 | (7.4 ± 1.9) × 105 | 2.1 ± 0.1 |
| NB10 | (1.1 ± 0.4) × 106 | (5.6 ± 1.3) × 106 | 4.9 ± 0.3 |
| M03 (rpoS null)b | 2.3 × 103 | 9.3 × 103 | |
Cells were grown for 16 h in LB20, washed and resuspended in either fresh LB20 or NSSM at 27°C for 180 min, and assayed for empA mRNA transcript number as described in the Materials and Methods. Values are means ± standard errors of the means.
Strain M03 is an rpoS mutant shown previously not to exhibit protease activity under any conditions assayed (2).
To reconcile the previously reported lack of protease activity in LB20-grown M93Sm cells (2, 3) with our detection of empA transcription, an experiment to measure both the transcription of empA and protease activity was performed. Sixteen-hour LB20 cultures of M93Sm and NB10 were washed and resuspended in LB20 as described above.
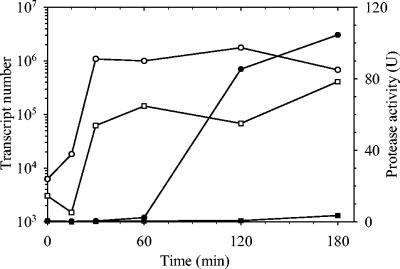
Samples for protease activity and RNA extraction were taken at 0, 15, 30, 60, 120, and 180 min. No protease activity was detected in the LB20-grown M93Sm culture. Protease activity was detected in the LB20-grown NB10 culture by 120 min (Fig. 1). Determination of empA transcription by qRT-PCR for both strains grown in LB20 revealed increases in message copy number from time 0 to 180 min of 108- and 134-fold for NB10 and M93Sm, respectively (Fig. 1). In both cases, increased empA expression was detected within 30 min.
FIG. 1.
Time series of protease activity (closed symbols) and empA mRNA expression (open symbols) using qRT-PCR in M93Sm (squares) and NB10 (circles) cells suspended in LB20 at ∼2 × 109 CFU ml−1. Assays for protease activity and RNA extractions were performed at 0, 15, 30, 60, 120, and 180 min. The amounts of empA transcripts per 10 ng of total RNA at time zero were 6.2 × 103 copies for NB10 and 3.0 × 103 copies for M93Sm.
The qRT-PCR data reported here contradicts previously reported Northern blotting data showing that M93Sm did not produce any empA mRNA in LB20 (2). We reran Northern blottings with 20 μg of total RNA per lane and detected a faint band in LB20-grown M93Sm cultures (data not shown). This demonstrated that the discrepancy was the result of the significantly lower sensitivity of Northern blot analysis compared to that of qRT-PCR. Comparison of qRT-PCR data with the new Northern blotting data showed that qRT-PCR detection is at least 2,000 times more sensitive than Northern blot analysis.
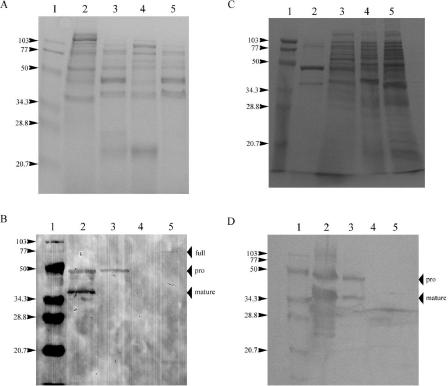
To determine whether the differences in EmpA protease activity between NB10 and M93Sm were due to differences in translation of message or posttranslational secretion and modification, Western blot analysis was performed. Cultures were grown to stationary phase (16 h) in both LB20 and NSSM. Proteins from culture supernatants and sonicated cell pellets were separated by SDS-PAGE. Examination of Coomassie blue-stained gels revealed numerous protein bands in both the supernatants and cell extracts under all conditions (Fig. 2A and C).
FIG. 2.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of EmpA secretion and activation. Shown are 16-h cultures of strains M93Sm and NB10 grown in either LB20 or NSSM. (A) Coomassie blue-stained proteins from LB20-grown cells. Lane 1, molecular size protein ladder; lane 2, supernatant from NB10 culture; lane 3, supernatant from M93Sm culture; lane 4, sonicated NB10 cells; lane 5, sonicated M93Sm cells. Numbers on the left indicate the mass (in kilodaltons) of the standard bands in lane 1. (B) Western blot of LB20 cultures, lanes, and standard labels the same as those in panel A. Position of the full peptide (full, 72 kDa), secreted pro-EmpA (pro, 48 kDa), and mature enzyme (mature, 38 kDa) are indicated with arrows on the right. (C) Coomassie blue-stained proteins from NSSM-grown cells. Lanes and standard labels are the same as those in panel A. (D) Western blot of NSSM cultures; lanes and standard labels are the same as those in panel A. Positions of the secreted pro-EmpA (pro, 48 kDa) and mature enzyme (mature, 38 kDa) are indicated with arrows on the right.
Western blotting revealed two anti-LasB-reactive bands in culture supernatants with estimated molecular masses of 48 and 38 kDa, representing pro-EmpA and mature EmpA, respectively. These two bands were in both LB20- and NSSM-grown NB10 supernatants (Fig. 2B and D, lanes 2). For M93Sm, both pro-EmpA and mature EmpA were present in NSSM supernatant, but only pro-EmpA was present in LB20 supernatant (Fig. 2B and D, lanes 3). In the lanes containing sonicated cell pellets, an approximately 72-kDa anti-LasB-reactive band was present in LB20-grown M93Sm cells (Fig. 2B, lane 5) but not in any of the cell samples from cultures with both mature EmpA and protease activity (Fig. 2B, lane 4, and D, lanes 4 and 5).
In the original report of the cloning of empA, Milton et al. (7) suggested that EmpA is synthesized as a preproenzyme. Sequence analysis indicated that removal of pre- and propeptides during secretion would result in a mature protein with a predicted molecular mass of 44.6 kDa. EmpA protease activity was associated with a 36-kDa protein, which suggested an additional processing step (7). Multiple processing steps have been proposed for other metalloproteases, including the hemagglutinin/protease of Vibrio cholerae (6). The data presented here support the suggestion of an additional processing step. In conditions where protease activity was detected in the supernatant, a ∼38-kDa band was detected with Western blotting. In all conditions with an active protease and for M93Sm grown in LB20, a ∼48-kDa band was also observed in the culture supernatant. The two bands at ∼48 and ∼38 kDa correspond to the size of the predicted secreted proenzyme and the mature protein, respectively (7).
In all cases where protease activity is detected, mature EmpA is present in the supernatant. Additionally, even when no protease activity is detected, stationary-phase M93Sm cells synthesize and secrete EmpA in the pro- form. This suggests that the difference in EmpA activity between M93Sm and NB10 occurs at the level of the activation of the proenzyme by the removal of a ∼10-kDa peptide and that this processing occurs after secretion. Further, the presence of stable pro-EmpA in the supernatant of LB20-grown M93Sm cells suggests that EmpA does not activate itself.
Salmon GI mucus acts to increase transcription of empA in both strains. However, EmpA activity is dependent upon successful secretion and processing of the nascent protein to an active mature form. These processes are linked to transcriptional regulation of empA in NB10, but they appear to be unlinked in M93Sm cells growing in LB20. Thus, we hypothesize that mucus initiates a process by which EmpA, secreted in a pro- form, is activated by a second proteolytic cleavage after secretion. The most likely mechanism by which this occurs is via the production and secretion of an additional protease that acts to remove the propeptide from EmpA and activate the enzyme. We are presently working to identify other proteases in V. anguillarum that may be involved in this process.
Acknowledgments
We thank David Laux for the gift of the anti-LasB antibodies.
The project was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant number 2002-35204-12252, awarded to D.R.N.
REFERENCES
- 1.Austin, B., and D. A. Austin. 1999. Bacterial fish pathogens: diseases in farmed and wild fish, 3rd ed. Springer, London, United Kingdom.
- 2.Denkin, S. M., and D. Nelson. 2004. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 70:4193-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denkin, S. M., and D. R. Nelson. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia, T., K. Otto, S. Kjelleberg, and D. Nelson. 1997. Growth of Vibrio anguillarum in salmon intestinal mucus. Appl. Environ. Microbiol. 63:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girouard, L., D. C. Laux, S. Jindal, and D. R. Nelson. 1993. Immune recognition of human Hsp60 by Lyme disease patient sera. Microb. Pathog. 14:287-297. [DOI] [PubMed] [Google Scholar]
- 6.Hase, C. C., and R. A. Finkelstein. 1991. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J. Bacteriol. 173:3311-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norqvist, A., B. Norrman, and H. Wolf-Watz. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 58:3731-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsson, J. C., A. Joborn, A. Westerdhal, L. Blomberg, S. Kjelleberg, and P. L. Conway. 1996. Is the turbot, Scophthalums maximus (L.), intestine a portal of entry for the fish pathogen Vibrio anguillarum? J. Fish Dis. 19:225-234. [Google Scholar]
- 10.O'Toole, R., J. von Hofsten, R. Rosqvist, P. E. Olsson, and H. Wolf-Watz. 2004. Visualization of zebrafish infection by FPF-labeled Vibrio anguillarum. Microb. Pathog. 37:41-46. [DOI] [PubMed] [Google Scholar]
- 11.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Windle, H., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]