Figure 2.
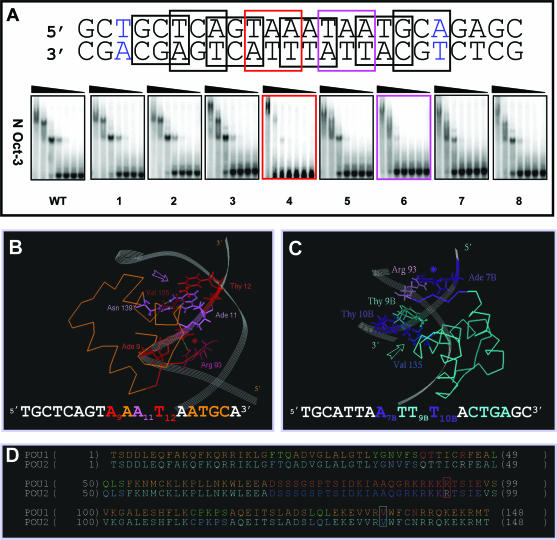
Structural determinants of the N-Oct-3 POU homodimerization on the AADC gene promoter (A) Mutation scanning of the 18 bp core region of the AADC fragment (the first and last base pairs are colored in blue) was carried out by selecting in turn 8 overlapping nucleotide triplets, in which A and G were substituted by C, and C and T substituted by G. The interactions between the N-Oct-3 DBD and the mutated AADC fragments were analyzed by EMSA and compared with the interaction with the non-mutated sequence (WT). The two most critical triplet mutations and their corresponding EMSA patterns have been highlighted. (B) Canonical interaction between the first monomer POUh and the optimal AAAT sub-site: anchoring into the DNA minor groove via the Arg 93/Ade 9 contact (red asterisk) and insertion of the recognition helix into the major groove via the Asn 139/Ade 11 and Val 135/Thy 12 contacts (pink arrow). (C) Interaction between the second monomer POUh and the ATTT sub-site: anchoring into the DNA minor groove via the Arg 93/Ade 7B contact (indigo asterisk) and insertion of the recognition helix into the major groove via the hydrophobic contacts between Val 135 and both Thy 9B and Thy 10B (turquoise arrow). (D) Location of Arg 93 (in red box), Val 135 (in turquoise box) and several important residues within the secondary structure elements of the first (POU1) and second (POU2) DBDs. Respective color coding is as follows: α-helices in brown and turquoise; POUh N-terminal extensions in red and indigo; linkers in dark brown and blue. POUs1 Gln 40, Thr41 and Arg 45 are colored in red, and POUs2 Thr 41 and Arg 45 in white.