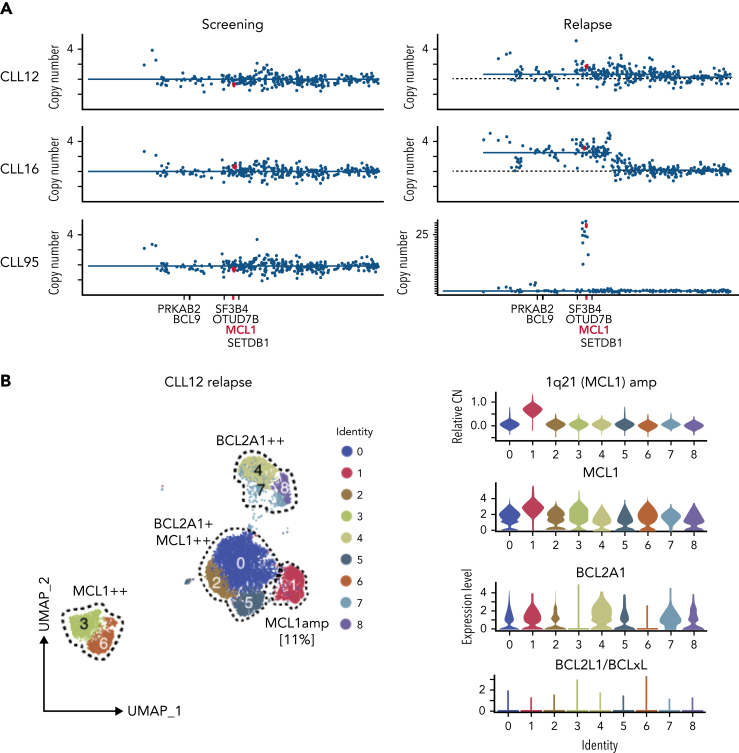
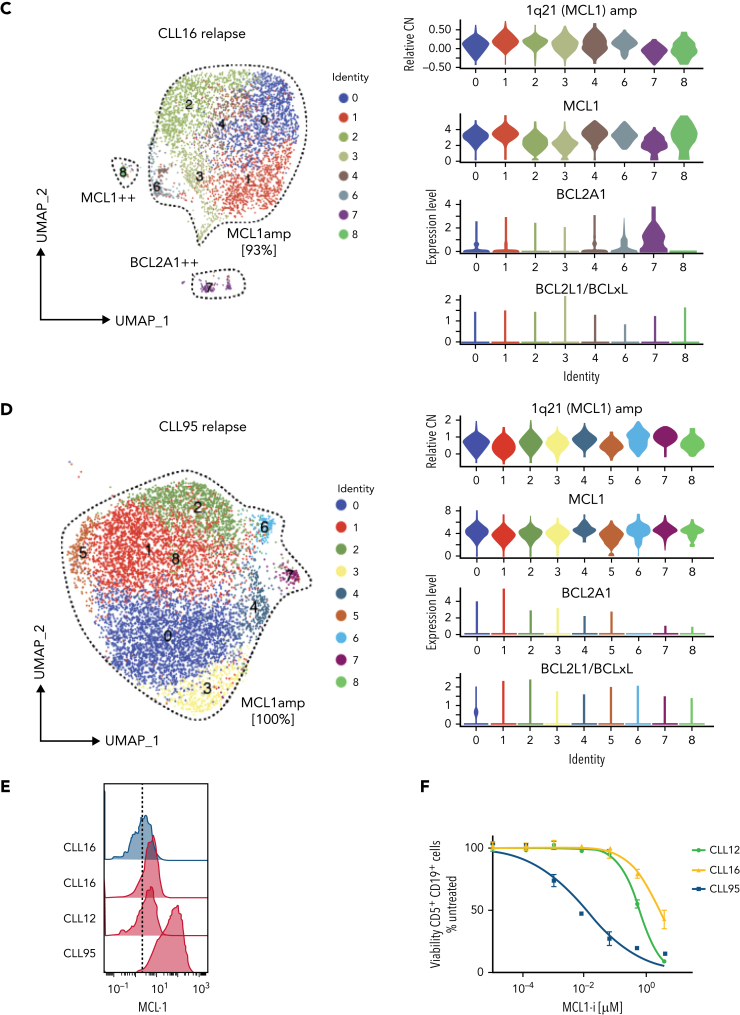
Figure 3.
Increased MCL1 expression at VEN relapse is only partially explained by amplification of the MCL1 gene. (A) Representation of the gained genomic regions in the 1q locus across the screening and relapsed samples from CLL12, CLL16, and CLL95 analyzed by WES. MCL1 is highlighted in red. Gray dotted lines indicate CN of 2. Blue lines indicate CN inferred from WES data. (B-D) UMAP projections (left) of CLL cells from (B) CLL12, (C) CLL16, and (D) CLL95 at relapse, with violin plots (right panels) showing inferred CN variation of 1q21 (MCL1) or relative expression of indicated prosurvival genes across the individual clusters. (E) Histograms showing mass cytometric analysis of MCL1 protein expression in viable (cisplatinlo) CD5+veCD19+ve PBMCs from screening samples from CLL16 (blue), relapsed samples from CLL16, CLL12, and CLL95 (all red). (F) PBMCs prepared at VEN-relapse were incubated in vitro for 24 hours with 0 to 4 μM S63845 (MCL1i). Viability (PI-ve) was measured in CLL cells (CD5+veCD19+ve), and data points represent the mean ± standard deviation of triplicate measurements in single experiments. CLL95 displayed marked sensitivity to MCL1i (LC50 10nM), while CLL12 and CLL16 had modest sensitivity (LC50s ∼600nM and ∼2700nM, respectively).