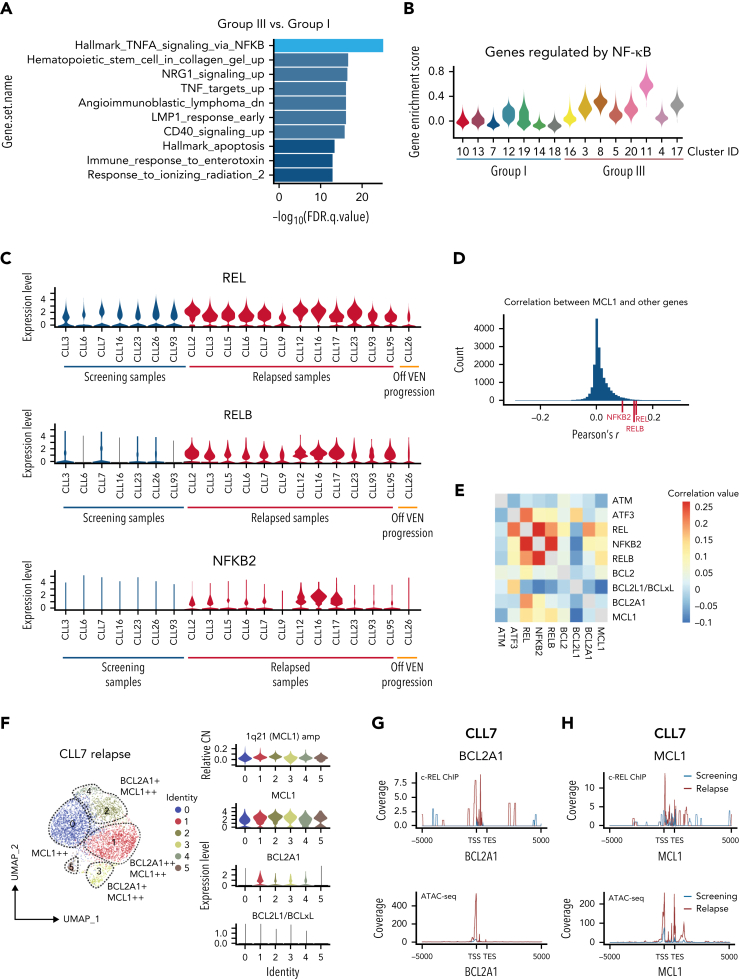
Figure 4.
Increased MCL1 expression driven by NF-κB at VEN relapse. (A) Bar plots showing pathway enrichment analysis in clusters from Group III (predominantly cells from VEN-relapsed samples) vs clusters in Group I (predominantly cells from screening samples). (B) Violin plots showing NF-κB pathway gene enrichment in Group III relapsed clusters. (C) Relative expression of REL, RELB, and NFKB2 in screening (n = 7; on left) or relapse (n = 13) samples; 1 patient (CLL26; on right) progressed while off VEN. (D) Pearson correlation coefficients were calculated between MCL1 and all other genes. The r score between MCL1 and REL, RELB, or NFKB2 are indicated with red ticks at the bottom. (E) Heatmap showing the correlation in expression between the indicated genes in the relapsed samples. Red shades indicate stronger positive correlations. (F) UMAP projections (left) of CLL cells from CLL7 at relapse, with violin plots (right panels) showing inferred CN variation of 1q21 (MCL1) or relative expression of indicated prosurvival genes across the individual clusters. (G-H) c-REL ChIP and chromatin accessibility at the BCL2A1 (G) and MCL1 (H) loci 5000 kb upstream of the transcriptional start site (TSS) or 5000 kb downstream of the transcriptional end site (TES) in purified CLL cells from screening (blue) or relapsed (red) samples from CLL7.