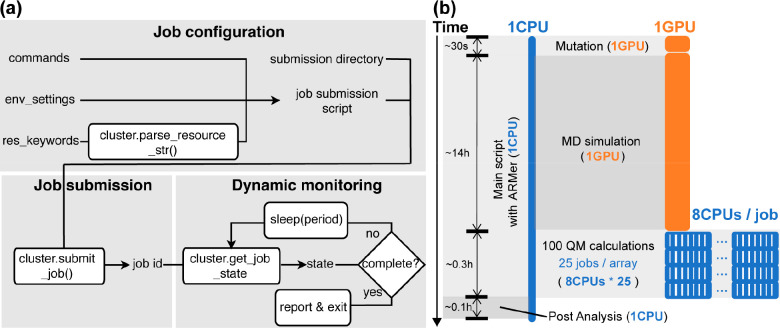
Figure 4.
Framework and application of adaptive resource manager (i.e., ARMer), a Python library used for adaptive computing resource allocation on high-performance computing cluster. (a) Variables and functions used by ARMer for configuration, submission, and dynamic monitoring of computational tasks. The variables and functions are encapsulated in a Job class. They can be called by a user to prepare a Python script that enables the construction of a high-throughput molecular modeling workflow with effective allocation of computing resources (e.g., CPU and GPU). (b) An exemplary application of ARMer to construct a workflow for high-throughput modeling of fluoroacetate dehalogenase (FAcD) mutants. In the workflow, a Python script that runs on a single-CPU thread leverages functions and variables from the Job class to manage the modeling subtasks (i.e., mutation, molecular dynamics, and quantum mechanics simulations) by configuring, submitting, and monitoring new job scripts. The MD job requests 1 GPU (in orange) and each QM job requests 8 CPUs (in blue). To submit and run individual QM calculations in parallel, a job array with a size of 25 is employed. The type of modeling subtasks, time usage, and resource cost are noted in the figure. Adapted with permission from ref (55). Copyright 2023 American Chemical Society.