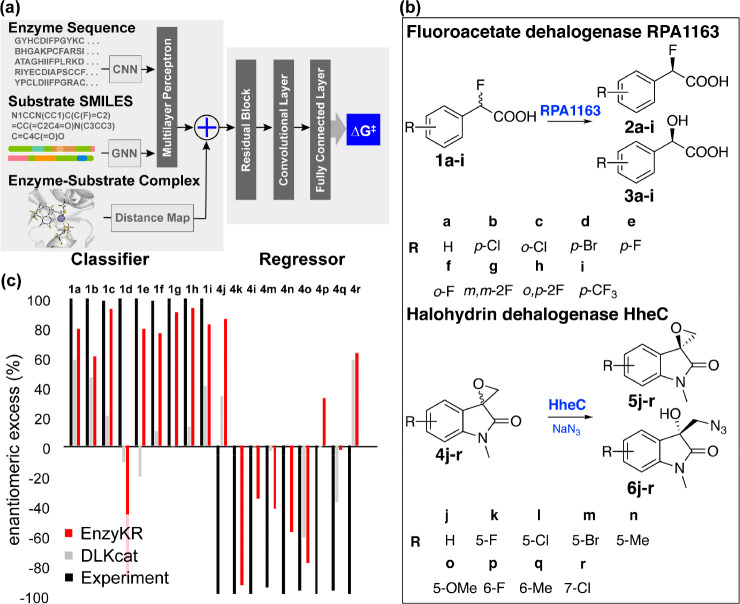
Figure 6.
Design and application of EnzyKR, a deep learning model for predicting the enantiomeric outcome of hydrolase-catalyzed kinetic resolution. (a) EnzyKR consists of a classifier and a regressor. Three types of input data for the classifier involve the enzyme–substrate complex structure, enzyme sequence, and simplified molecular-input line-entry system (SMILES) string. The distance map derived from the complex structure is encoded using a 2D convolutional neural network (CNN). The multiple sequence alignments (MSA) of the enzyme sequences are also encoded by a 2D CNN model. The substrate SMILES strings are encoded by a graph neural network (GNN) model. The embeddings from the classifier and the interaction maps are used as input for the regressor. The regressor involves one module of cross-attention, followed by residual blocks consisting of three 2D dilated convolution layers, one 2D batch norm layer, and one ReLU layer. Two layers of a fully connected neural network (i.e., multiple-layer perceptron) are employed to conduct regression between the extracted feature and the activation free energy. (b) The test reactions used to assess the ability of EnzyKR to predict the outcomes of kinetic resolution. The test set involves 18 enantioselective hydrolytic reactions catalyzed by two hydrolases. RPA1163 is a fluoroacetate dehalogenase that catalyzes the C–F bond hydrolysis in 9 fluoroacetic acid derivatives labeled from a to i. HheC is a halohydrin dehalogenase that catalyzes the stereoselective epoxide ring-opening in 9 spiro-epoxyoxindoles derivatives labeled from j to r. (c) The predicted enantiomeric excess (ee%) values of EnzyKR (red) and the baseline model DLKcat (gray) for 18 enantiomer pairs in hydrolase-catalyzed reactions. The experimental ee% value is shown in black. A positive ee% value indicates that the S-configuration is favored.