Abstract
Aims:
Aortic stenosis (AS) is a degenerative valve condition that is under-diagnosed and undertreated. Detection of AS using limited 2D echocardiography could enable screening and improve appropriate referral and treatment of this condition. We aimed to develop methods for automated detection of AS from limited imaging datasets.
Methods:
Convolutional neural networks were trained, validated, and tested using limited 2D transthoracic echocardiogram (TTE) datasets. Networks were developed to accomplish two sequential tasks; 1) view identification and 2) study-level grade of AS. Balanced accuracy and area under the receiver operator curve (AUROC) were the performance metrics used.
Results:
Annotated images from 577 patients were included. Neural networks were trained on data from 338 patients (average N = 10,253 labeled images), validated on 119 patients (average N = 3,505 labeled images), and performance was assessed on a test sets of 120 patients (average N = 3,511 labeled images). Fully automated screening for AS was achieved with AUROC 0.96. Networks can identify no significant (no, mild, mild/moderate) AS from significant (moderate, or severe) AS with an AUROC = 0.86 and between early (mild or mild/moderate AS) and significant (moderate or severe) AS with an AUROC of 0.75. External validation of these networks in a cohort of 8502 outpatient TTEs showed that screening for AS can be achieved using parasternal long-axis imaging only with an AUROC of 0.91.
Conclusion:
Fully-automated detection of AS using limited 2D datasets is achievable using modern neural networks. These methods lay the groundwork for a novel method for screening for AS.
Keywords: aortic stenosis, screening, machine learning, echocardiography, transthoracic echocardiography
Introduction
Aortic stenosis (AS) is an enormous public health problem that affects over 12.6 million adults and worldwide causes an estimated 102,700 deaths annually.1 Recently, there has been interest in earlier identification of AS and evidence that many patients may not be appropriately treated.2,3 These observations motivate the study of novel methods to identify AS. Here, we evaluate whether machine learning (ML) methods can accurately identify AS using limited 2D imaging datasets that are well suited for disease screening.
Little is known about how to improve identification and treatment of AS. Employing a population-based comprehensive transthoracic echocardiogram (TTE) screening approach would be prohibitively expensive. Automated interpretation of limited echocardiography datasets is an attractive alternative approach to disease detection, especially with the rise of point of care ultrasound (POCUS) devices. Barriers to automating AS detection relate to the complex nature of this diagnosis, the need to integrate information across multiple images for any given study, and datasets that are not routinely annotated as part of routine clinical care.
Classically, accurate grading of AS relies on integration of numerous structural and hemodynamic parameters from across multiple imaging planes.4 From the perspective of disease screening, certain features of AS such as valve thickness and calcium burden are readily apparent on 2D images. While deep learning methods can now surpass humans in certain medical image classification tasks5,6, common classifier designs take only individual images as input and applications in echocardiography have so far focused only on viewpoint identification, image segmentation, and assessments of ventricular function and myocardial diseases.7–10 ML approaches to AS so far are limited to using echocardiogram reports (thereby requiring expert image interpretation to work)11,12 or are limited to very small numbers13 or focusing only on severe disease.14 None have focused on assessing the continuum of AS severity using limited images with the goal of establishing tools suitable for automated disease screening. Here we develop methods that can produce a coherent single diagnosis (severity of AS) from limited 2D datasets.
Methods
Echocardiograms
This work was approved by the Tufts Medical Center IRB. The echocardiograms originate from TTEs performed between 2011–2020 at a high-volume tertiary care center (Tufts Medical Center, Boston, MA). The echocardiograms were acquired as part of routine clinical care. The CardioVascular Imaging Center is Intersocietal Accreditation Commission (IAC) accredited and is equipped with ultrasound units from major vendors (Philips®, Toshiba®, Siemens®). By using standardized digital imaging and communications in medicine (DICOM) images, these methods are intended to be vendor-independent. Echocardiograms were included based on the presence or absence of AS. Images were not selected for inclusion based on image quality. Patients with prior aortic valve replacements were excluded. Other cardiac findings including other concomitant structural heart disease and rhythm abnormalities (i.e. atrial fibrillation) were not excluded.
Image acquisition and preprocessing
Images were acquired by trained sonographers with methods consistent with current American Society of Echocardiography (ASE) guidelines.15 For this study, we used metadata to identify and discard all spectral Doppler, color-flow Doppler, and M-mode recordings, keeping only 2D cardiac transthoracic echocardiogram (TTE) images. To minimize compute time and enhance transportability and to position these networks for use in future screening environments, the first frame of each PLAX or PSAX AoV loop was automatically selected for use in the prediction models. If there were multiple PLAX or PSAX AoV acquisitions in a study (as is often the case) predictions used the first frame from each acquisition to arrive at study-level AS prediction. All images were standardized to 112×112 pixel resolution. Consistent with routine clinical care, there are no view or diagnostic label annotations available for images when they are collected.
De-identification
Leveraging known region locations that are encoded within the DICOM storage format, propriety software was created to automatically identify the image burn region that contains protected health information (PHI). By excluding these imaging regions from the data copy, images were reliably de-identified. A 10% samples of the included de-identified images was manually reviewed to confirm no PHI was included.
Limited View labels
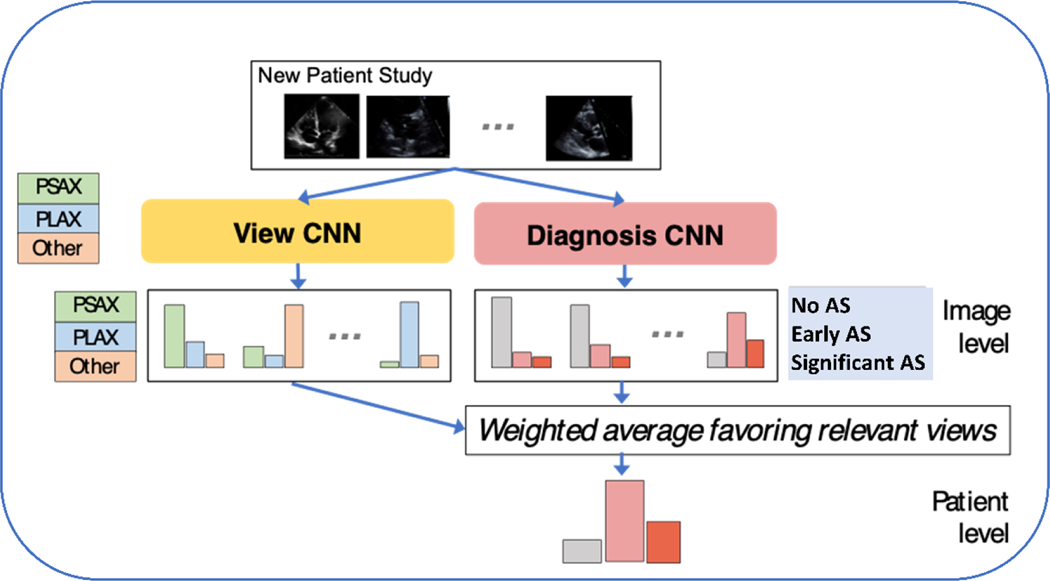
The study setup followed the cognitive steps involved in diagnosing AS by echocardiography (Figure 1), specifically view recognition followed by view interpretation. We collected expert annotations of a limited number of view types with two goals in mind; 1) to evaluate (and validate) automated view classification networks and 2) prioritized views are used in subsequent AS diagnostic models. Labels were assigned to examples of the parasternal long axis (PLAX), parasternal short axis at the level of the aortic valve (PSAX AoV). These views were purposely selected because they are standard views that can visualize the AoV and can be prioritized in a limited screening environment. For evaluation of view the classification tasks apical 2-chamber (A2C), and apical 4-chamber (A4C) views were also labeled. An ‘Other’ super-category label that covered other 2D views was also collected. Doppler imaging was not included in this study because these acquisitions are not routinely collected during point-of-care ultrasound (POCUS) imaging studies and because Doppler image acquisition requires a high level of skill that is often only available in dedicated echocardiogram laboratories.
Figure 1. Approach to Automated Identification of Aortic Stenosis.
Convolutional neural networks (CNN) were trained and tested to identify view type and AS diagnostic category using limited 2D datasets.
An echocardiogram annotation tool was built to facilitate view annotation (Supplemental Figure 1). Annotators (board certified echocardiographers or American Registry for Diagnostic Medical Sonography credentialed sonographers) assigned labels to ≥ 2 examples of each imaging view for each of the 599 studies included in our labeled set. Agreement between labelers was assessed on a set of 50 echocardiograms that were labeled in duplicate (Supplemental Table 1, Supplemental Table 2).
Diagnostic labels
The presence or absence of AS and the grade of AS (if present) were assigned by a cardiologist with specialty training in echocardiography. AS classification was assigned during clinical care in standard fashion following an integrative approach as recommended by current guidelines (i.e. integrating information across all available images of all view types for a given patient).4 The reference grade of AS for this study was taken directly from the clinical imaging report. AS labels for these experiments are shown in Table 1. Echocardiograms representing the full spectrum of AS pathologies were purposely included. To focus this work on potential automated screening use cases, and with recognition that inter-reader agreement of disease severity is modest,16 we grouped standard severity levels into 3 diagnostic classes: “no AS”, “early AS” (combining mild and mild-to-moderate), and “significant AS” (combining moderate and severe). In a screening environment (upstream of the traditional echocardiogram laboratory), the primary clinical question is which individuals should be referred for comprehensive echocardiography and AS-related care.
Table 1. Aortic Stenosis Reference Labels.
AS Reference Labels. Aortic Stenosis severity was assigned using an integrative approach consistent with current American Society of Echocardiography Guidelines.4 The Severe AS reference label includes both high gradient and low gradient subtypes. AS severity was pulled from the echocardiogram report as assigned by the clinical reader. As part of routine care an additional label of ‘Mild to Moderate’ AS was assigned when hemodynamic profiles overlap the ‘Mild’ and ‘Moderate’ severity classes. This label was preserved for these experiments. Valve area represents the continuity equation derived valve area. LVOT is the left ventricular outflow tract diameter. mmHg is millimeters of mercury, m/s is meters per second.
| Reference AS Severity | Grading Thresholds |
|---|---|
| Severe | valve area of < 1.0 cm2, peak velocity ≥ 4.0 m/s, or mean gradient ≥ 40 mmHg |
| valve area of < 1.0 cm2, peak velocity < 4.0 m/s or mean gradient < 40 mmHg and LVOT derived stroke volume ≤ 35 mL/m2 | |
| Moderate | valve area of 1.0–1.5 cm2, peak velocity 3.0–4.0 m/s, or mean gradient 20–40 mmHg |
| Mild | valve area of >1.5 cm2, peak velocity 2.6–2.9 m/s, or mean gradient <20 mmHg |
Datasets and Experimental Design
Our experiments focused on assessing performance on echocardiogram studies from never-before-seen patients. These experiments were done in a manner consistent with Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME) checklist.17 The PRIME checklist for this study is available upon request.
Our final dataset consisted of 599 fully-labeled TTE studies, where each study has a diagnosis label (no AS, early AS, or significant AS) as well as some images with view labels. We have released this dataset to researchers worldwide as the Tufts Medical Echocardiogram Dataset, version 2 (TMED-2). Most patients contributed only one study, but multiple studies from a small number of patients (22 out of 577) were included to improve dataset size. Each patient’s data were assigned to exactly one set to properly assess generalization across individuals. The labeled data were divided into training (60%), validation (20%), and test sets (20%). We ensured that the ratio of diagnostic classes was the same across training, validation, and test (~21% no AS, ~29% early AS, and ~50% significant AS). Dataset composition by label is summarized in Table 2 (for diagnosis task) and Supplemental Table 3 (for view task).
Table 2: AS Diagnosis Label Cohorts across Train/Test Splits.
AS Diagnosis Label Cohorts across Train/Test Splits in TMED-2. We show the number of echocardiogram studies assigned to train/valid/test sets across all 3 possible aortic stenosis (AS) severity levels for the fully-labeled dataset of 599 studies. Image counts represent the mean over 3 splits, as the exact number of images per study differs across splits. No split deviates more than 12% from the reported mean here. Each patient’s data were assigned to exactly one set to properly assess generalizability to new patients, while preserving similar proportions of each AS severity level across train and test.
| Number of Studies | Number of Labeled Images | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | None | Early AS | Sig AS | Total | None | Early AS | Sig AS | |
| Train* | 360 | 76 | 103 | 181 | 10253 | 999 | 1316 | 7938 |
| Valid | 119 | 25 | 34 | 60 | 3505 | 344 | 402 | 2759 |
| Test | 120 | 26 | 34 | 60 | 3511 | 339 | 408 | 2764 |
indicates that the training set also included an additional set with only view labels but no AS diagnosis label. This view-only set contained 705 studies representing 7694 labeled images, see Supplement Table 3.
Abbreviations: None represents no AS, Early AS represents mild and mild/mod AS, Sig AS means moderate, moderate/severe, and severe AS.
Each ML method was allowed to fit parameters to the training set, select hyperparameters based on performance on the validation set, and report results on the test set. To improve the reliability of our results, we repeated the process of training a model and evaluating its performance across 3 separate, independent random partitions of all data into training, validation, and test sets. We report average performance across these 3 test sets. In addition to using the full training and validation set (479 studies), we also considered two levels of reduction (165 and 56 studies, roughly 33% and 11% of the full size). The same full-size test sets (120 studies) were always used to compare final performance.
Deep Neural Nets for View and Diagnosis Classification
We trained two neural networks: one view classifier and one diagnosis classifier. Each used the same backbone neural network architecture: a wide residual network with 28 layers containing 5,931,683 parameters18. Each network takes one image as input and produces a predicted probability vector. We discuss how we aggregate predictions across images in the next paragraph. The view classifier is trained to produce a 5-way probabilistic view classification (PLAX, PSAX AoV, A4C, A2C, or Other) given a single image. To train, we minimize 5-class cross entropy summed over all view-labeled images in the labeled set. The diagnosis classifier is trained so the same network produces two separate outputs given a single image: the primary output is a 3-way probabilistic vector indicating the diagnosis (no, early, or significant AS), and the auxiliary output is a 5-way probabilistic vector indicating the view type. We use multi-task training, where the loss function is a sum of the 3-class diagnosis cross entropy and 5-class view cross entropy, summed over all view-labeled images. We found this multi-task training delivered better diagnosis performance. After multi-task training, only the 3-class diagnosis output is used (the separately trained view neural network is a better view classifier than the auxiliary output). Each model was trained via stochastic gradient descent until the validation balanced accuracy for its primary task did not improve for at least 30 epochs. The actual epochs needed vary from 150-1000 depending on the method used.
Producing One Study-Level Diagnosis from Many Images
Given the trained image-to-view and image-to-diagnosis classifier networks described above, our goal was to automate assignment of a study-level AS severity diagnosis: one vector summarizing the holistic interpretation of all images in a study. To accomplish this, we applied an approach which we call Prioritized View weighting19, which we developed on an earlier, smaller dataset. The intuitive motivation is that diagnostic predictions made from images that show the aortic valve (PLAX or PSAX AoV views) should be considered stronger evidence than predictions of disease severity from other view types. Concretely, our Prioritized View procedure obtains a study-level probabilistic prediction in three steps. First, using the image-to-diagnosis classifier to produce a 3-class probability vector indicating AS severity for every image in the study. Second, use the image-to-view classifier to predict the probability of a relevant view (PLAX or PSAX) for every image. Finally, compute a weighted average over the 3-class vectors from step one, weighting each by the probability from step two. We compare this Prioritized View approach to an alternative Simple Average that treats diagnoses from all images equally without any weighting by the view classifier.
Performance Metrics
Throughout the evaluation of both view and diagnostic tasks we use balanced accuracy as the performance metric of interest. Standard accuracy does not adequately assess performance when the data has imbalanced class distributions, as seen in both view and diagnostic tasks. Balanced accuracy is computed in two steps: compute the fraction of true members of each class that are correctly recognized, then average this fraction across all classes.
To further assess our method’s utility as a screening tool, we use receiver operating curve (ROC) analysis and report area-under-the-curve for several potential use cases: (A) distinguishing no AS from any AS (early and significant), (B) distinguishing early AS from significant AS, and (C) distinguishing non-significant (no AS or early AS) from significant (moderate, moderate/severe, or severe) AS.
External Validations
External validation of the view classifer was done using the The Stanford EchoNet Dynamic dataset.20 This dataset contains 10,030 images of the A4C view type, gathered using completely different patient populations, clinical teams, and label assignments than our Tufts-focused dataset. Since all 10,030 images are A4C views, we report our view classifier’s accuracy on this dataset. We used all available A4C images in this dataset, which are provided at 112×112 resolution (the same resolution we used for our images).
Two external validation studies of the AS diagnostic classifiers were done. The first was a temporal external validation performed on TTEs done at Tufts Medical Center from May to July, 2022. These studies represent consecutive clinically indicated TTEs with an AS classification that was independently reviewed for this study (no AS, early AS, significant AS, defined in an identical fashion to the model derivation tasks).
Next, we performed an external validation study on data provided by iCardio.ai. The data consisted of TTEs performed between 2018–2020 by an outpatient diagnostic imaging company. For this validation we had access to a single imaging view (PLAX) and the AS diagnostic label for the study. This validation was designed to test model performance on limited 2D acquisitions. The TTEs for this cohort were performed for independent medical practices and clinics in over 13 states in the US and data were acquired with ultrasound units from 4 major vendors (GE®, Philips®, Teratech®, Acuson®). AS grade was assigned by a COCATS level III echocardiographer.
Statistical Analysis
To evaluate the classification performance of each neural network, we report the balanced accuracy on the test set. To assess binary discrimination between two classes, we also report the area under the ROC curve. For each performance metric, we report a 95% confidence interval computed using 5,000 bootstrapped samples of the test set. We average this reporting across 3 independent partitions or “splits” of the data into training and test.
Results
The clinical characteristics of the patients included in this study are shown in Table 3a. The primary labeled cohort included 577 patients. The median age was 74 (IQR 63–82). 43% of the patients were women. 86% of the study population was Caucasian. The hemodynamic parameters of the echocardiograms are shown in Table 3b. The median aortic valve peak velocity was 2.89 m/s (2.29–3.67), the median peak gradient was 34.6 mmHg (21.0–54.0 mmHg), and the average mean gradient was 18.1 mmHg (11.9–30.6 mmHg). The average LV ejection fraction was 60% (55–65%).
Table 3a. Baseline Characteristics of Patients and Echocardiograms in TMED-2 Dataset.
All baseline characteristics refer to the entire cohort of 577 patients. All values are medians unless otherwise specified. IQR is interquartile range. BP is blood pressure, PCI is percutaneous coronary intervention. CABG is coronary artery bypass grafting, CVA is cerebrovascular accident.
| Patient Characteristics (N = 577) | |
|---|---|
| Age | 74 (IQR 63–82) |
| Sex (% women) | 43% |
| Race | 85% Caucasian 4% Black 3% Latino 8% other |
| Height (inches) | 66 (63–69) |
| Weight (lbs) | 174 (146–208) |
| BMI (kg/m2) | 27.8 (24.2–32.1) |
| Systolic BP (mmHg) | 129 (116–144) |
| Diastolic BP (mmHg) | 72 (63–79) |
| Hypertension (%) | 80% |
| Hyperlipidemia | 68% |
| Congestive Heart Failure | 33% |
| Diabetes | 31% |
| Prior myocardial infarction | 13% |
| Prior PCI | 17% |
| Prior CABG | 13% |
| Prior CVA | 10% |
| Current smoking | 8% |
Table 3b. Echocardiograms in TMED-2 Dataset.
Hemodynamic values were extracted from the medical record. Screening tasks correspond to the automated diagnostic tasks studied: 1) Any AS (vs none), 2) Early AS vs Significant AS, 3) No significant AS (vs significant AS). Not all values are available for every study, the number reporting each value are shown in the left hand column. All values are median +/− interquartile range (IQR) unless otherwise specified. AS is aortic stenosis. LV is left ventricle, V2 max is the peak continuous wave velocity. AV is aortic valve. LV ejection fraction was assessed by integrating the biplane method of disks summation (modified Simpson’s rule) with overall visual assessment. Max gradient is the maximum continuous wave gradient across the aortic valve. Mean gradient is the mean continuous wave gradient across the aortic valve. mmHg is millimeters of mercury.
| Echocardiogram Study Characteristics | ||||||
|---|---|---|---|---|---|---|
| Screening Task | All | No AS | Early AS | Significant AS | ||
| AS Grade | No AS | Mild AS | Mild/moderate AS | Moderate AS | Severe AS | |
| N | 599 | 127 (21.2%) | 144 (24.0%) | 27 (4.5%) | 132 (22.0%) | 169 (28.2%) |
| Stroke Volume, mL (n = 566) | 59.6 (46–75.6) | 54 (41–70.8) | 61 (47–83.1) | 50 (43.0–69.7) | 66.0 (52.0–83.0) | 59.0 (47.8–72.0) |
| LV ejection fraction, % (n = 599) | 60 (55–65) | 55 (45–60) | 60 (55–65) | 60 (55–65) | 60 (55–65) | 60 (55–65) |
| V2 max, m/s (n = 507) | 2.89 (2.29–3.65) | 1.73 (1.28–2.00) | 2.45 (2.32–2.64) | 2.89 (2.80–2.97) | 3.26 (3.11–3.47) | 4.32 (3.96–4.65) |
| AV max gradient mmHg (n = 508) | 34.4 (21–52.7) | 12.1 (6.8–16.1) | 24.1 (21.8–28.1) | 34.1 (32.1–35.2) | 42.6 (38.9–48.4) | 74.4 (63.0–87.0) |
| AV mean gradient, mmHg (n = 491) | 18.1 (11.9–30.0) | 6.7 (4.1–8.5) | 13.2 (11.6–15.1) | 17.8 (15.8–19.4) | 23.0 (20.2–26.2) | 42.0 (35.0–50.0) |
Partitioning of the dataset is shown in Table 2. The fully-labeled set contains both diagnosis and view labels (599 studies representing 577 patients, 43823 total images of which 17270 have view labels). After preprocessing, the median study in our dataset contains 70 images (5th-95th percentile range 48–105, min-max range 15–181). The median number of images with view labels is 19 (5–95th range 4.9–71, min-max range 1–107). Fully-labeled data were divided into training (60%, 360 studies), validation (20%, 119 studies), and test sets (20%, 120 studies).
View classification
Our view classifiers deliver 97% balanced accuracy on test set when averaged over the 3 partitions of TMED-2. Balanced accuracy for the view task increases notably as the size of the available training and validation data increases from 90.3% with only 56 studies (95% bootstrap CI 87.5–90.4%) to all 97.0% with all 476 studies (95% bootstrap CI 95.9–97.5%, Supplementary Table 4). Using power-law curve-fitting that has been empirically successful at characterizing deep learning performance as dataset sizes increase21, we project that labeled-set-only balanced accuracy could improve to 98.5% if 1000 labeled studies were available for model training and validation (Supplemental Figure 2).
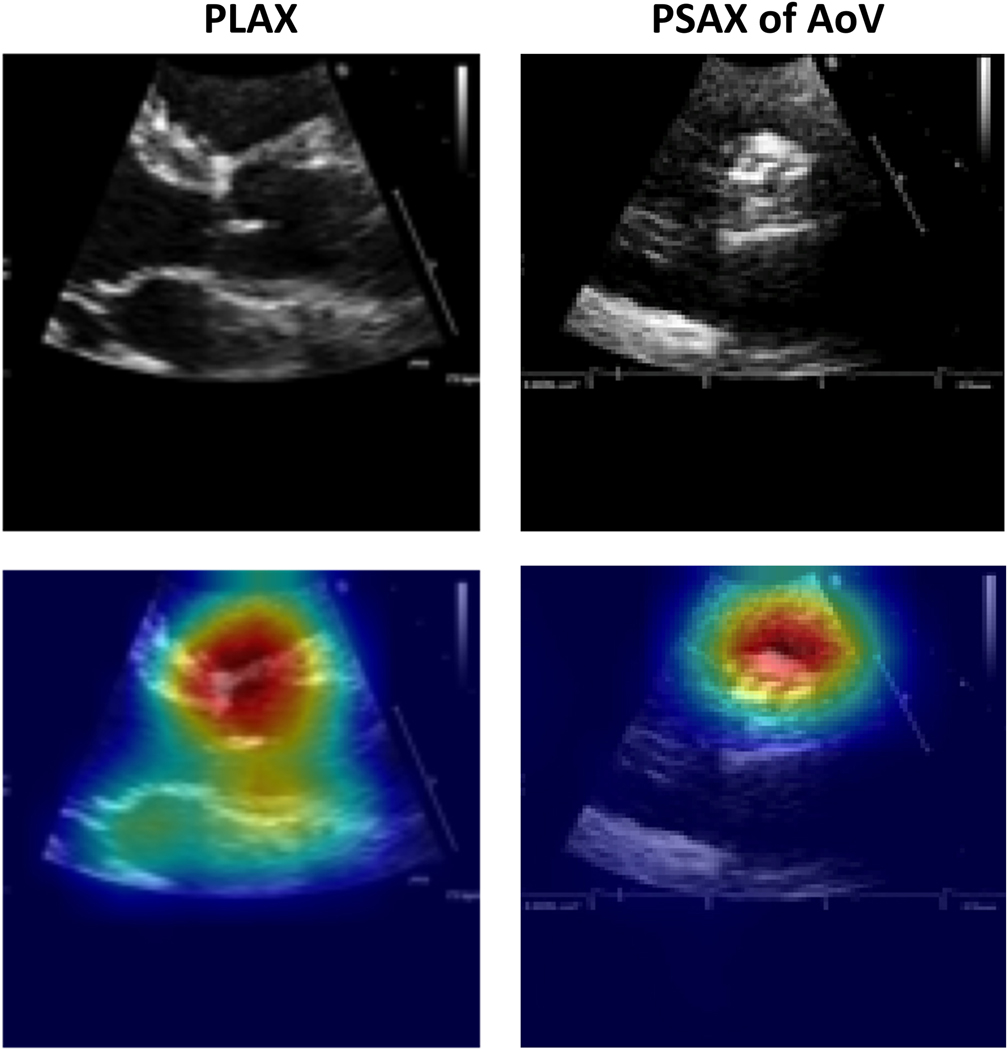
To sanity-check prediction quality, we used Grad-CAM22 to generate visual explanation heat-maps for our view classifier on select images from our test set (Figure 2). These visuals suggest that view predictions depend on relevant regions of the aortic root and aortic valve instead of irrelevant background data.
Figure 2.
Grad-CAM visualizations of view predictions. Examples of PLAX (left) and PSAX AoV-level (right) views in our test set and their Grad-CAM visualizations. The original image is shown at the top, and the corresponding Grad-CAM visualization is shown below (original image with heat map overlay). The model correctly predicted the images to be PLAX and PSAX views, respectively, and correctly focused on the relevant region of the heart for making the predictions. The hotter the color, the more important the pixel in making the class discriminative decisions.
External validation of view classification
Accuracy at recognizing A4C views from the external EchoNet dataset is 93.4% (95% bootstrap CI: 93.2–93.8%) averaged over 3 splits on the full labeled set (476 studies for training and validation, 120 for test). When the available labeled data are smaller, performance is naturally less accurate: 81.1% when developed on 165 studies and 66.1% when developed on 56 studies (Supplement Table 5).
Diagnosis performance using limited 2D images related to a patient
Supplementary Figure 2 shows how study-level diagnosis classification improves with more labeled data across two strategies for averaging across all images to make a coherent study-level diagnosis (Prioritized View vs. Simple Average). On our full dataset, the Prioritized View strategy delivers 74.5% balanced accuracy for the 3-way AS diagnosis task compared to 34.9% for Simple Average (Supplementary Table 6). Note that random guessing baseline would achieve 33%.
On the largest training split, the multi-task training for our diagnosis classifier took ~14 hours on a Nvidia® RTX6000 Graphics Processing Unit (GPU). Using an already-trained network, it takes approximately 0.4 seconds to get an AS diagnostic prediction for a typical study.
Using automatic study-level diagnosis as a preliminary screening tool for AS
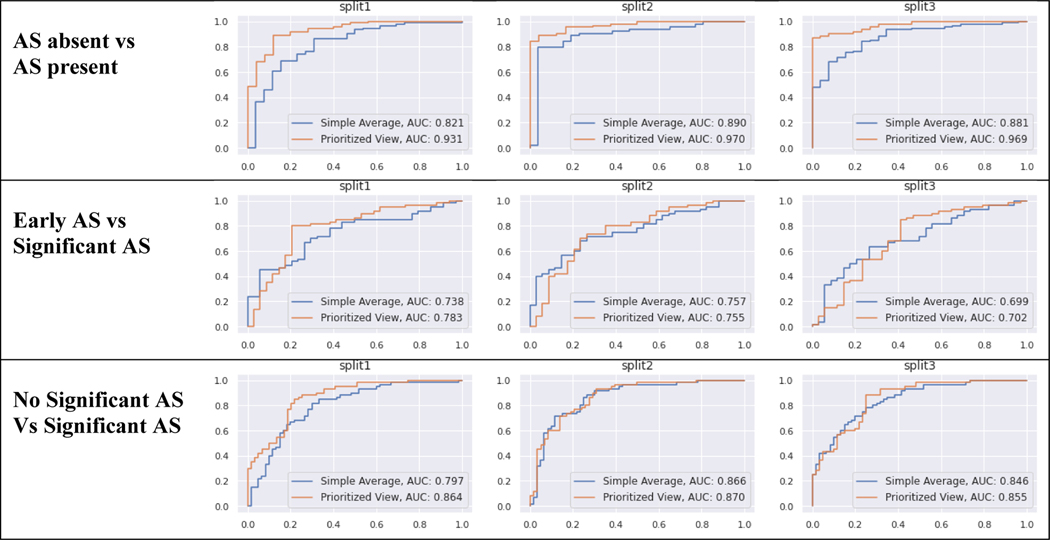
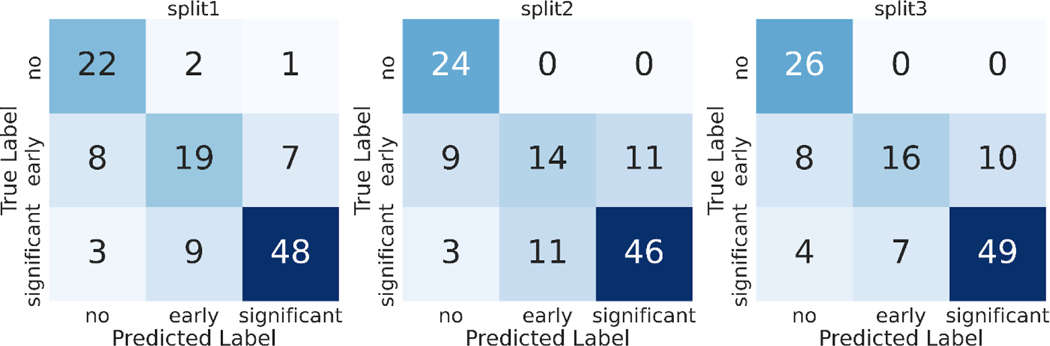
Discriminatory performance for various clinical use cases are shown in Table 4, Figure 3. A) Using limited 2D images, AUROC = 0.96 for screening for any AS, B) AUROC = 0.75 for identifying early (mild or mild/moderate) AS vs significant (moderate, moderate/severe, severe) AS, and C) AUROC = 0.86 for no significant AS (no AS and early AS) vs significant (moderate, moderate/severe, or severe) AS. Using these data, our methods demonstrate sensitivity of 88.3% and specificity of 88% for detecting AS. The confusion matrices for these predictions are shown the Figure 4.
Table 4. Model Discrimination (AUROC) for AS Screening Tasks.
Model Discrimination for three binary screening tasks: 1) AS absent vs AS present, 2) Early AS vs. Significant AS, 3) no significant AS vs Significant AS. We report the Area under the Receiver Operator Curves (AUROC) for each task, averaged over 3 random training-validation-test splits of the data. The 95% bootstrap CI of this average is in parentheses. Methods two methods of aggregating image-level predictions to a study-level diagnosis: simple averaging or a weighted averaged that prioritizes specific views (PLAX or PSAX) that depict the aortic valve and are thus relevant for AS diagnosis.
| Model | AS absent vs AS present | Early AS vs Sig AS | Sig AS vs No Sig AS |
|---|---|---|---|
| Simple Average | 0.86 (0.81 – 0.91) | 0.73 (0.67 – 0.79) | 0.84 (0.79 – 0.88) |
| Prioritized View | 0.96 (0.93 – 0.97) | 0.75 (0.68 – 0.81) | 0.86 (0.82 – 0.90) |
Figure 3. Diagnostic Classification Receiver Operator Curves.
Each set of experiments was run with 3 random training-validation-test splits of the data (labeled split1, split2, split3). The top row represents screening for AS: AS absent vs AS present (any severity). Middle row represents early AS (mild, mild/moderate) vs significant AS (moderate, severe). Bottom row represents non-significant AS (none, mild, mild/moderate) vs significant AS (moderate, severe). Each line gives the performance of one prediction strategy for aggregating across all images in a study: Prioritized View and Simple Average. Each column shows the results for one partition of the TMED-2 data into training/test.
Figure 4: Confusion Matrices for AS Severity classification.
Each set of experiments was run with 3 random training-validation-test splits of the data (labeled split1, split2, split3). We report the test set confusion matrix from classifiers trained on each of the 3 train/test splits of our TMED-2 dataset.
External validation of AS classification
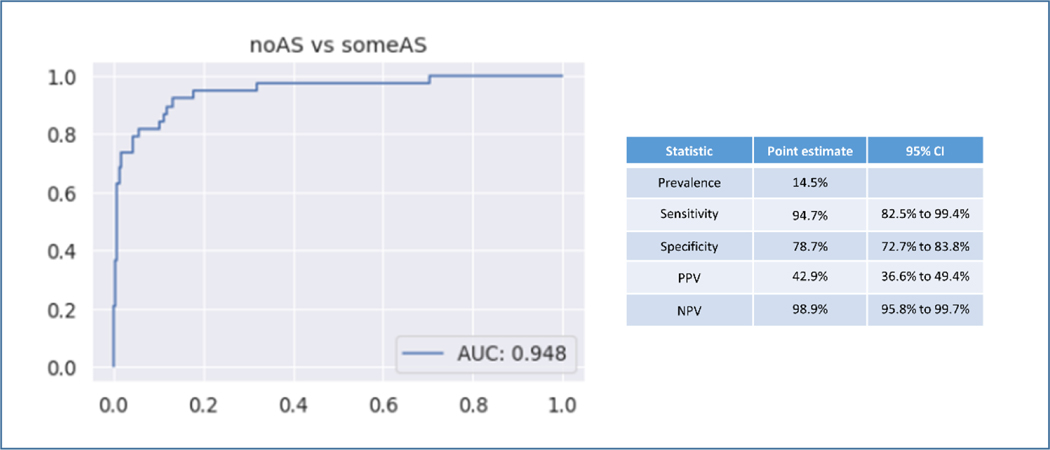
In the temporal validation the AS classifier was used to study 263 consecutive TTEs acquired at Tufts Medical Center with independently verified AS grade as assigned by a board certified echocardiographer. For the diagnostic screening task of identifying AS (all grades) the AUROC was 0.95. In this external validation cohort the prevalence of AS was 14.5%. The sensitivity was 94.7% and the specificity was 78.7%. Positive predictive value (PPV) was 42.9% and negative predictive value (NPV) was 98.9% (Figure 5). For the task of identifying significant AS (moderate and severe) vs no significant AS (no AS, mild, mild-moderate AS) the AUROC was 0.95 (Supplemental Figure 3).
Figure 5. Temporal Validation of Diagnostic Classifier.
Temporal External Validation of the fully automated network for AS identification. AUROC shown on the left for 263 consecutive TTEs at Tufts Medical Center. AS diagnosis was independently reviewed for this study.
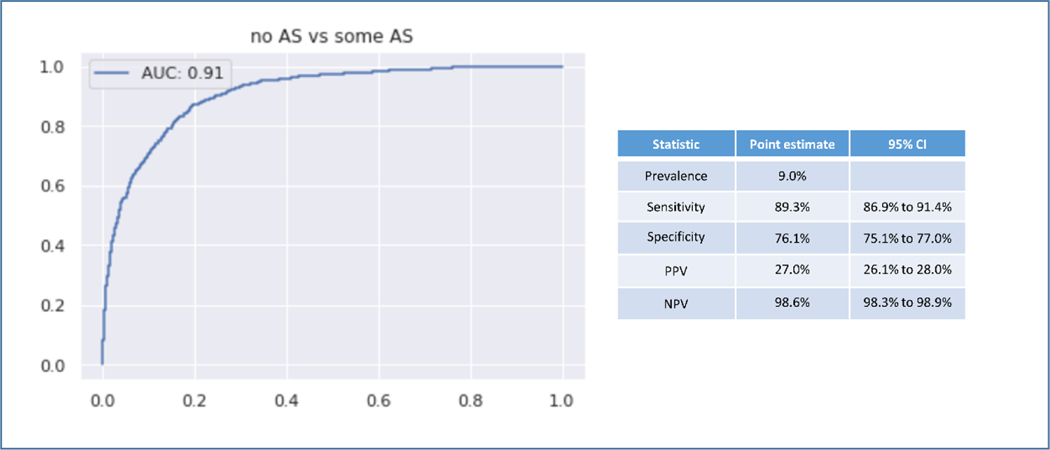
In the fully external validation using a single PLAX view, the AS classifier was used to screen for AS in 8502 echocardiograms. For the screening task the AUROC was 0.91. In this cohort the prevalence of AS was 9.0%. The sensitivity was 89.3% and the specificity was 76.1%. The PPV was 27.0% and the NPV was 98.6% (Figure 6).
Figure 6. Fully External Validation.
Fully External Validation on 8502 echocardiograms from iCardio.ai. This validation study used only the PLAX view. AS diagnosis was assigned by fully independent echocardiographers.
Discussion
Novel approaches to AS case identification are needed in order improve treatment rates for this condition. Here we develop methods for fully automated detection of AS from limited TTE datasets. We show that automated detection of AS is possible using modern deep learning classifiers and that these networks are generalizable across different datasets. These tools can broadly characterize the presence or absence of AS and the severity of disease and are well suited for identifying patients who should be referred for comprehensive echocardiography. These results represent important steps toward establishing a novel approach to AS case identification.
These models are not designed to comprehensively phenotype AS as can be done with complete TTE. Instead, we view this work as a method to move case identification upstream of the echocardiogram laboratory. With an estimated incidence rate of severe AS of 4.4%/year in the general population >65 years of age, it is clear that many patients go unrecognized.23 The sensitivity and specificity of contemporary care with cardiac auscultation for detecting significant valve disease is only 44% and 69% respectively.24 Performance of auscultation is likely to be even lower for detecting more mild disease where murmurs are less intense. An automated screening program that uses limited 2D datasets—embedded within or upstream of hospital or clinic-based echocardiogram laboratories—might improve case identification and referral. While the discriminatory performance of these models appears excellent, the positive predictive value is modest. This is related in part to the relatively low prevalence of significant AS and should be viewed in the context of a very high negative predictive value (i.e. very few cases will be missed).
These tools could enable studies to address the profound treatment disparities for patients with severe AS25,26 or interrogate emerging evidence that many patients with severe AS are not treated.3,27,28 Automated screening could allow for large scale studies of the natural history of AS and also uncover potential biases in the care pathway of patients with this condition. Certainly, additional studies are needed to assess whether automation tools that enable effective screening and timely referral can improve outcomes for patients with AS. Automated detection of AS might also enable studies of early interventions to halt disease progression.29 Classically, it has been challenging to study early stage disease since early AS is asymptomatic. Fully automated interpretation of limited echocardiography may be worthwhile if effective treatments emerge, or for enabling trial recruitment for treatment of earlier stage disease. The methods presented here do not use Doppler images and so are potentially suited to use with POCUS devices.
The modern networks studied here are attractive for the field of echocardiography because they can learn competitive models from small labeled datasets. These models used a single frame from the cine loops. Use of the time-varying feature sets almost certainly contains additional information, however this added information has to be balanced against the computational requirements needed to process more complex datasets. This network was designed to be scalable and require the least-information necessary to be clinically valuable. Additionally, as demonstrated with the external validation study, these networks can ingest complete or partial studies and assign a diagnostic label. This flexibility positions these methods for use with limited acquisitions in screening environments. Here study-level diagnoses were achieved using a novel view-prioritized approach which uses a view-classifier to identify views deemed relevant for the diagnostic task of interest (here AS). Diagnostic classifier predictions from these relevant views are then prioritized via a weighted average to predict a coherent study-level diagnostic label. We present validation studies of the sequential view and diagnostic tasks to emphasize the tiered approach used here that we believe can be applied to automate other complex imaging diagnoses.
The data used in these studies are released as part of our Tufts Medical Echocardiogram Database version 2 (TMED-2, data and code available at https://tmed.cs.tufts.edu/). TMED-2 substantially increases the number of publically released studies, increases resolution to 112×112 from 64×64, and increases available view types compared to our smaller earlier release.19 This database covers a range of AS pathologies and will support the development of novel methods to automate screening for complex imaging diagnoses. The notable accuracy gains possible on external data with 3x increases in dataset size illustrate the critical need for efforts to make labeled datasets available to researchers worldwide.
There are a few limitations to this work that must be recognized. The presented echocardiograms come from a single academic center and diagnostic labels were assigned as part of routine clinical care. Non-white patients were under-represented in this cohort though the echocardiogram-based imaging diagnosis of AS should not have any biologic differences based on race. This study did not include outcome data or information from other imaging modalities to confirm disease severity. While the number of labeled echocardiograms is modest, these networks are notable in that they can learn from small labeled sets. This is important for future model development where labels are expensive and time consuming. While more complex low flow/ low gradient subtypes may be misclassified, we minimize this risk by collapsing moderate and severe AS into a single ‘significant AS’ category that should be referred for comprehensive study and care. This is by design and is important for future screening trials. Prior efforts that focus only on high flow/ high gradient subtypes14 would miss a significant number of cases that represent severe disease with lower flow profiles. With release of our code and images, we encourage additional external validations of our work. We expect performance would improve with higher image resolutions, larger neural networks, or use of all frames from cineloops rather than the first frame only; we kept resolutions modest (112 × 112 pixels) and used only one frame to achieve a tractable balance between accuracy and training time. On modern GPUs each neural network we trained already requires dozens of hours on the largest version of our dataset.
Conclusion
ML approaches optimized for echocardiography can successfully identify AS using limited 2D datasets. These methods lay the groundwork for fully automated screening for this disease and future study of interventions to improve outcomes.
Supplementary Material
Highlights.
Automated detection of aortic stenosis (AS) is a novel approach to diagnosis.
ML methods were trained to detect AS from limited 2D echo images
Fully-automated screening for AS using limited datasets is achievable.
Release of a TTE database will encourage collaboration.
Acknowledgements:
This work was supported by the National Institutes of Health Tufts CTSI NIH CTSA UL1TR002544). BW received funding from the National Institutes of Health (K23 AG055667)
Funding:
This work is supported by NCATS grant UL1TR002544
BW is supported by NIH grant K23AG055667
Abbreviations:
- AS
aortic stenosis
- TTE
transthoracic echocardiography
- ML
machine learning
- VHD
valvular heart disease
- AVR
aortic valve replacement
- SSL
semi-supervised learning
- DICOM
digital imaging and communications in medicine
- PLAX
parasternal long axis
- PSAX AoV
parasternal short axis at the level of the aortic valve
- TMED-2
Tufts Medical Echocardiogram Dataset, version 2
Footnotes
Disclosures:
Declaration of interest: BW has done consulting work with iCardio.ai and US2.ai unrelated to the present work and is co-founder of CVAI Solutions. CVAI Solutions created the software for the de-identification procedures though currently has no related commercial pursuits.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yadgir S, Johnson CO, Aboyans V, et al. Global, Regional, and National Burden of Calcific Aortic Valve and Degenerative Mitral Valve Diseases, 1990–2017. Circulation. 2020;1670–80. [DOI] [PubMed] [Google Scholar]
- 2.Lindman BR, Sukul D, Dweck MR, et al. Evaluating Medical Therapy for Calcific Aortic Stenosis: JACC State-of-the-Art Review. J Am Coll Cardiol. 2021;78(23):2354–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li SX, Patel NK, Flannery LD, et al. Trends in Utilization of Aortic Valve Replacement for Severe Aortic Stenosis. J Am Coll Cardiol. 2022. Mar 8;79(9):864–77. [DOI] [PubMed] [Google Scholar]
- 4.Baumgartner H, Hung J, Bermejo J, et al. Recommendations on the Echocardiographic Assessment of Aortic Valve Stenosis: A Focused Update from the European Association of Cardiovascular Imaging and the American Society of Echocardiography. J Am Soc Echocardiogr. 2017. Apr;30(4):372–92. [DOI] [PubMed] [Google Scholar]
- 5.Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316(22):2402–10. [DOI] [PubMed] [Google Scholar]
- 6.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Gajjala S, Agrawal P, et al. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation. 2018. Oct 16;138(16):1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang D, He B, Ghorbani A, et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature. 2020. Apr 9;580(7802):252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy G, Cheng PP, Yuan N, et al. High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy With Cardiovascular Deep Learning. JAMA Cardiol. 2022. Feb 23; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tromp J, Seekings PJ, Hung CL, et al. Automated interpretation of systolic and diastolic function on the echocardiogram: a multicohort study. Lancet Digit Heal. 2022. Jan;4(1):e46–54. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta PP, Shrestha S, Kagiyama N, et al. A Machine-Learning Framework to Identify Distinct Phenotypes of Aortic Stenosis Severity. JACC Cardiovasc Imaging. 2021. Sep;14(9):1707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Playford D, Bordin E, Mohamad R, et al. Enhanced Diagnosis of Severe Aortic Stenosis Using Artificial Intelligence: A Proof-of-Concept Study of 530,871 Echocardiograms. JACC Cardiovasc Imaging. 2020. Apr;13(4):1087–90. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Ojha BD, Aranoff ND, et al. Classification of aortic stenosis using conventional machine learning and deep learning methods based on multi-dimensional cardio-mechanical signals. Sci Rep. 2020. Dec 16;10(1):17521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W, Nazzari H, Namasivayam M, et al. Identifying Aortic Stenosis With a Single Parasternal Long-Axis Video Using Deep Learning. J Am Soc Echocardiogr. 2022. Oct 30; [DOI] [PubMed] [Google Scholar]
- 15.Mitchell C, Rahko PS, Blauwet LA, et al. Guidelines for Performing a Comprehensive Transthoracic Echocardiographic Examination in Adults: Recommendations from the American Society of Echocardiography. J Am Soc Echocardiogr. 2019;32(1):1–64. [DOI] [PubMed] [Google Scholar]
- 16.Haji K, Wong C, Neil C, et al. Multi Reader Assessment of Accuracy and Interobserver Variability in Aortic Stenosis by Echocardiography. Hear Lung Circ. 2019;28:S258. [Google Scholar]
- 17.Sengupta PP, Shrestha S, Berthon B, et al. Proposed Requirements for Cardiovascular Imaging-Related Machine Learning Evaluation (PRIME): A Checklist. JACC Cardiovasc Imaging. 2020. Sep;13(9):2017–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zagoruyko S, Komodakis N. Wide Residual Networks. In: Procedings of the British Machine Vision Conference 2016. 2016. p. 87.1–87.12. [Google Scholar]
- 19.Huang Z, Long G, Wessler B, et al. A New Semi-supervised Learning Benchmark for Classifying View and Diagnosing Aortic Stenosis from Echocardiograms. Proc Mach Learn Healthc Conf. 2021. Jul 30; [Google Scholar]
- 20.Ghorbani A, Ouyang D, Abid A, et al. Deep learning interpretation of echocardiograms. npj Digit Med. 2020. Dec 24;3(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hestness J, Narang S, Ardalani N, et al. Deep Learning Scaling is Predictable, Empirically. ArXiv. 2017. Dec 1; [Google Scholar]
- 22.Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In: 2017 IEEE International Conference on Computer Vision (ICCV). IEEE; 2017. p. 618–26. [Google Scholar]
- 23.Durko AP, Osnabrugge RL, Van Mieghem NM, et al. Annual number of candidates for transcatheter aortic valve implantation per country: current estimates and future projections. Eur Heart J. 2018;39(28):2635–42. [DOI] [PubMed] [Google Scholar]
- 24.Gardezi SKM, Myerson SG, Chambers J, et al. Cardiac auscultation poorly predicts the presence of valvular heart disease in asymptomatic primary care patients. Heart. 2018;104(22):1832–5. [DOI] [PubMed] [Google Scholar]
- 25.Batchelor W, Anwaruddin S, Ross L, et al. Aortic Valve Stenosis Treatment Disparities in the Underserved: JACC Council Perspectives. J Am Coll Cardiol. 2019;74(18):2313–21. [DOI] [PubMed] [Google Scholar]
- 26.Clark KA, Chouairi F, Kay B, et al. Trends in transcatheter and surgical aortic valve replacement in the United States, 2008–2018. Am Heart J. 2022. Jan;243:87–91. [DOI] [PubMed] [Google Scholar]
- 27.Tang L, Gössl M, Ahmed A, et al. Contemporary reasons and clinical outcomes for patients with severe, symptomatic aortic stenosis not undergoing aortic valve replacement. Circ Cardiovasc Interv. 2018;11(12):1–12. [DOI] [PubMed] [Google Scholar]
- 28.Brennan JM, Bryant A, Boero I, et al. PROVIDER-LEVEL VARIABILITY IN THE TREATMENT OF PATIENTS WITH SEVERE SYMPTOMATIC AORTIC VALVE STENOSIS. J Am Coll Cardiol. 2019. Mar;73(9):1949. [Google Scholar]
- 29.Lindman BR, Merryman WD. Unloading the Stenotic Path to Identifying Medical Therapy for Calcific Aortic Valve Disease. Circulation. 2021. Apr 13;143(15):1455–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.