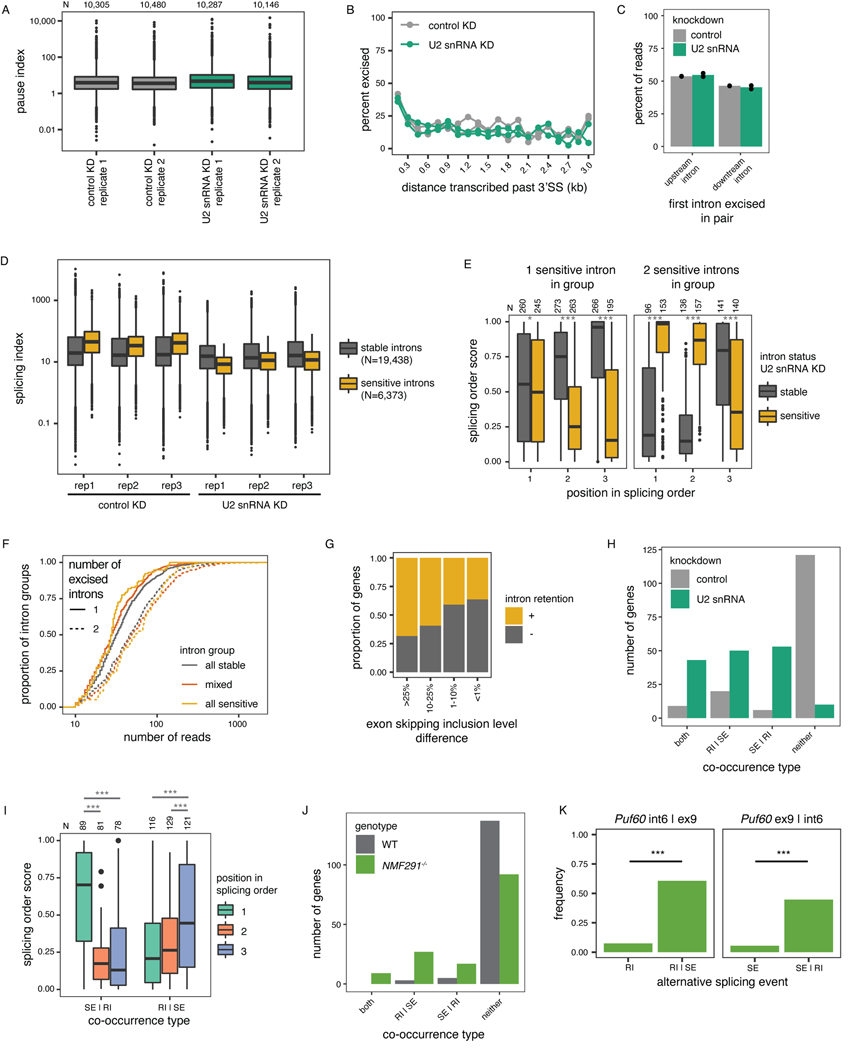
Extended Data Figure 7.
(related to Figure 5). A) NET-seq pause index upon control or U2 snRNA KD. B) Percent of excised introns as a function of the distance transcribed past the 3’ SS for nano-COP reads from control and U2 snRNA KD. Two biological replicates per condition are displayed. C) Splicing order in nano-COP data from control and U2 snRNA KD for pairs of consecutive introns. Dots represent biological replicates. D) Total RNA-seq splicing index for introns that are sensitive to U2 snRNA KD compared to those that are stable. Three biological replicates (“rep”) per condition are shown. E) Summed splicing order score for introns that are sensitive or stable upon U2 snRNA knockdown as a function of their position in splicing orders in WT HeLa cells. F) Number of reads mapping to each intron group as a function of the number of excised introns and their status upon U2 snRNA KD. G) Proportion of genes with and without intron retention in short-read RNA-seq from upon U2 snRNA KD, as a function of the exon skipping level difference between control and U2 snRNA KD in the same genes. H) Number of genes showing co-occurrence of skipped exons (SE) and retained introns (RI) in dRNA-seq, as depicted in Fig. 5F. I) Summed splicing order score for RIs upon U2 snRNA knockdown as a function of their position in splicing orders in WT HeLa cells. Intron groups are composed of one intron involved in RI and two introns involved in SE. J) Number of genes showing co-occurrence of SE(s) and RI(s) in cDNA-PCR nanopore sequencing data from WT and NMF291−/− (U2 snRNA mutant) mice53. K) Example of a SE and RI co-occurrence in Puf60 in NMF291−/− mice. ***: p-value < 0.001 in the one-sided binomial test assessing co-occurrence of SE and RI. In A), D), E) and I), boxplots elements are shown as follows: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers. In E) and I), groups were compared using a two-sided Wilcoxon rank-sum test; *: p-value < 0.05; ***: p-value < 0.001.