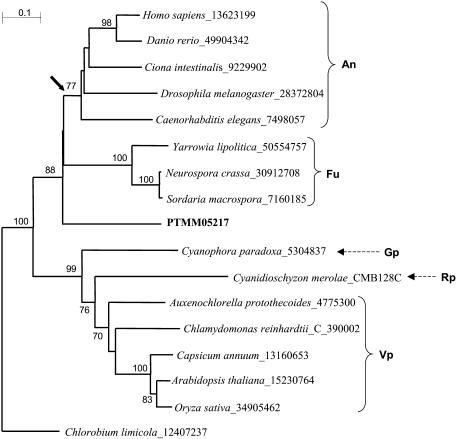
Figure 4.
Phylogenetic analysis of putative ATP:citrate lyases (ACLs) from P. tricornutum. The figure shows a neighbor-joining tree of putative or described ACLs representing the major lineages in which ACL-like genes are known, including the most highly conserved ACL-like sequence in P. tricornutum (in bold). The sequences were trimmed to a conserved core of 185 amino acids. The GenBank GI sequence identifiers of the proteins used are shown following the species name. The C. reinhardtii and C. merolae sequences were obtained from their respective genome browsers rather than from GenBank and their predicted gene identification numbers are given. Bootstrap values above 70% (of 1,000 replicates) are shown. Members of the Animalia (An), Fungi (Fu), Glaucocystophyta (Gp), Rhodophyta (Rp), and Viridiplantae (Vp) are indicated. The Chlorobium limicola ACL is the sole prokaryotic ortholog known to date and was used to root the tree. The arrow indicates the time of fusion of α- and β-ACLs into a single gene as proposed by Fatland et al. (2002). Scale bar = 0.1 substitutions/site.