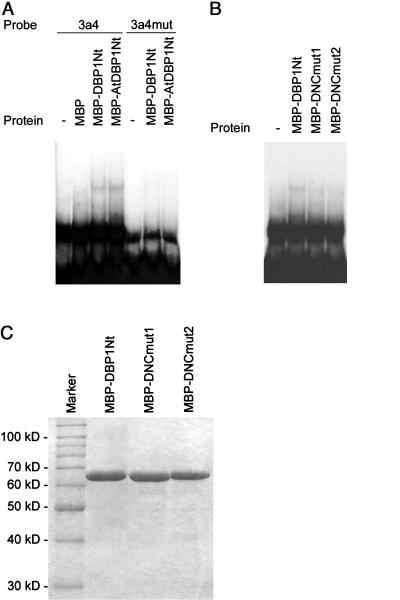
Figure 3.
A, DNA binding in vitro by the N-terminal regions of DBP1 and AtDBP1. Recombinant MBP fusion proteins were expressed in E. coli and purified by amylose affinity chromatography, according to the manufacturer's instructions (New England Biolabs, Beverly, MA). Electrophoretic mobility shift assays were performed as described above. The sequences of the oligonucleotides used to generate the double-stranded mutated probe 3a4mut are the following, with changed nucleotides shown in bold: 3a4mut-plus, 5′-TCGACTAGGCGGCAGGAGTTTGGGAGAGTCTCCCTC-3′; and 3a4mut-minus, 5′-TCGAGAGGGAGACTCTCCCAAACTCCTGCCGCCTAG-3′. Both 3a4 and 3a4mut probes were radioactively labeled as described above. Labeling was quantified by liquid scintillation and found to be almost identical in both cases. The same amount of each labeled probe was used in EMSA. B, Effect of mutations in the DNC motif on DNA binding by the N-terminal region of DBP1. Mutations in MBP-DNCmut1 and MBP-DNCmut2 were introduced using the megaprimer method (Datta, 1995). Mutant proteins were expressed in E. coli and purified as described above. Purified recombinant proteins were incubated at a final concentration of 1 μm with a radiolabeled 3a4 DNA probe (0.3 nm) for 10 min at room temperature in binding buffer [20 mm HEPES-KOH, pH 7.6, 4 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 10% (v/v) glycerol], prior to electrophoresis on a native 6% (w/v) polyacrylamide gel in 0.5× Tris-borate/EDTA buffer. C, SDS-PAGE of purified recombinant proteins used in EMSA.