Figure 2.
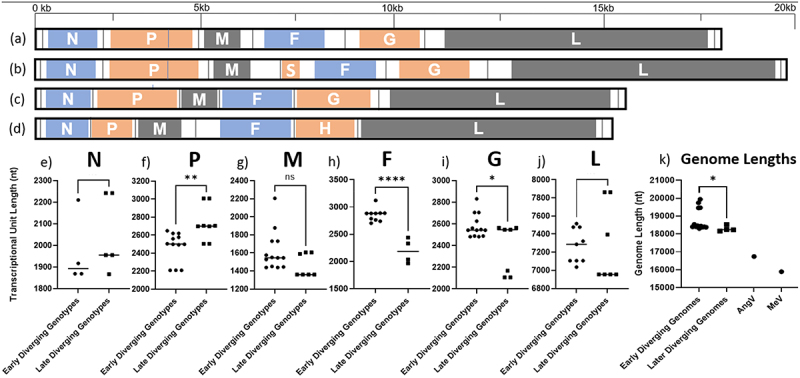
Genomic organization of henipaviruses and related species: of the selected genomes (a) the HeV genome has the conserved gene order N-P-M-F-G-L where each ORF is encoded on a separate mRNA which differs from (b) which is the MeliV genome characteristic of LayV, MojV, GAKV, DARV, and related viruses where the F gene encodes an additional ORF X in the 5’ UTR producing the S protein. AngV (c) has the smallest genome and each mRNA is likely monocistronic. MeV (d) which is representative of the morbillivirus genus as an outgroup is markedly smaller compared to (a) and (b). The transcriptional unit length (all nucleotides from last intergenic residue to the first residue of the next intergenic region) from each major branch of the henipavirus phylogeny compared to each other for (e) the N gene of genomes with intergenic region present in the leader, (f) for P gene in all genomes, (g) M in all genomes, (h) F transcriptional unit (including ORF X/S protein CDS for early diverging genomes), (i) G gene, (j) for the L gene in all genomes with intergenic sequence in the trailer, and (k) total reported genome length. The length of early diverging henipavirus genomes (including LayV and MojV) is longer compared to later diverging genomes (including HeV, NiV, and CedV) and the transcriptional unit encoding the F protein and/or S protein is significantly longer (f).
