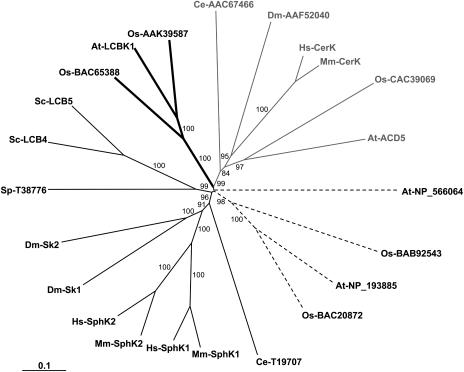
Figure 8.
Phylogenetic tree of SphKs and CerKs. Deduced amino acid sequences were aligned using ClustalW multiple alignment and visually refined using BioEdit Sequence Alignment Editor 4.8.8. The phylogenetic tree was generated by the neighbor-joining method using the PAUP 4.0b10 package. Branch lengths are scaled proportional to genetic distance. Bootstrap values (percent) were obtained with 1,000 replicates and those exceeding 50% are indicated above the supported branches. Sequences from the following organisms were used: At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe. Known SphKs and CerKs are as follows: At-LCBK1 (accession no. NP_568432), At-ACD5 (accession no. AAQ62904), Dm-Sk1 (accession no. AAF48045), Dm-Sk2 (accession no. AAF47706), Hs-SphK1 (accession no. AAF73423), Hs-SphK2 (accession no. AAF74124), Hs-CerK (accession no. BAC01154), Mm-SphK1 (accession no. AAC61697), Mm-SphK2 (accession no. AAF74125), Mm-CerK (accession no. BAC01155), Sc-LCB4 (accession no. NP_014814), Sc-LCB5 (accession no. NP_013361). Accession numbers of putative SphKs, CerKs, and long-chain-base kinases are indicated in the dendrogram. Thin shading, thick shading, gray, and dotted lines refer to Cluster SphKs, long-chain-base kinases, CerKs, and putative long-chain-base kinases, respectively.