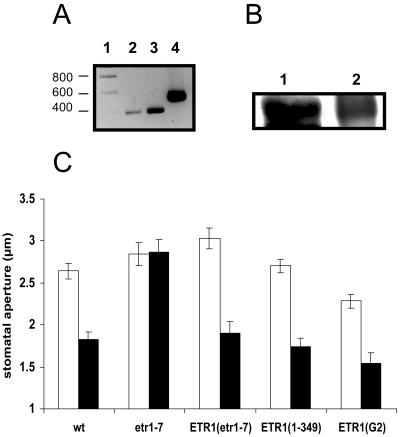
Figure 2.
ETR1 is expressed in wild-type Arabidopsis guard cells and is required for H2O2-induced closure. A, RT-PCR of RNA extracted from guard cells (lane 2) and whole leaves (lane 3). Lane 4, Genomic DNA positive control; lane 1, DNA marker (indicated in basepairs). For RNA extractions, frozen leaf material was blended in a Waring blender (3 × 15 s) in water (2.5 g: 20 mL containing 1.5% TRIzol reagent [Invitrogen, Paisley, UK]) and ice, and the homogenate filtered through a 100-μm mesh (Spectramesh, VWR International, Poole, UK). This was repeated a further four times in the same mixture. The guard cell-enriched epidermal fragments (>95% guard cells, as assessed by FDA/DAPI staining; Hey et al., 1997) were then homogenized in TRIzol reagent (1 mL) with glass beads in a Fastprep bead beater (Fisher, Loughborough, UK) to break open the guard cells. RNA was extracted following the TRIzol RNA extraction procedure provided by the manufacturers. Reverse transcription was performed on DNAsed RNA, with PCR primers designed against sequences unique to ETR1; forward primer was ETR1F (5′-GTTTGTGAATCTGATGGAGGG-3′) and reverse primer was ETR1R (5′-GTTGTTTTGTGAATTTCTCG-3′). Genomic DNA was used as a control for the PCR to check that the RT products were from cDNA, and the PCR product subsequently sequenced. B, Western blot of guard cell proteins. Lane 1, Guard cell proteins; lane 2, yeast proteins. Guard cell-enriched epidermal fragments were prepared as above (but minus TRIzol) and the proteins extracted in extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm DTT, 10 mm Na3VO4, 10 mm NaF, 50 mm α-glycerophosphate, 1 mm PMSF, 5 μg mL−1 aprotinin, and 5 μg mL−1 leupeptin) in the Fastprep bead beater, centrifuged for 2 × 20 min at 15,500g at 4°C, and the supernatant concentrated using Microcon (VWR International) spin columns. Yeast proteins were isolated from cells expressing full-length ETR1, as described (Rodriguez et al., 1999). Proteins were prepared for SDS-PAGE by incubating at 37°C for 1 h in SDS buffer without DTT and electrophoresed on a 7.5% SDS-polyacrylamide gel. Western blotting was performed using an anti-ETR1 (C terminus) antibody (Insight Biotechnology) and detected using enhanced chemiluminescence (GE Healthcare, Bucks, UK). C, The loss-of-function etr1-7 mutant is insensitive to H2O2. Arabidopsis leaves were floated for 3 h under continuous illumination (200–250 μE m−2 s−1) in MES/KCl buffer (5 mm KCl/10 mm MES/50 μm CaCl2, pH 6.15). Once the stomata were fully open, leaves were treated with H2O2 for a further 3 h. The leaves were subsequently homogenized individually in a Waring blender for 30 s and the epidermal fragments collected on a 100-μm nylon mesh (SpectraMesh). Stomatal apertures from epidermal fragments were then measured using a calibrated light microscope attached to an imaging system (Leica QWin software, Leica, Milton Keynes, UK). Stomatal closure response to H2O2 (100 μm) in wild type (wt); etr1-7; etr1-7 complemented with full-length ETR1 (ETR1[etr1-7]); etr1-7 complemented with ETR1 truncated at 349 (ETR1[1-349]); etr1-7 complemented with ETR1 containing a mutation in the G2 box of the HK domain (ETR1[G2]). White bars, control; black bars, H2O2. Data are expressed as mean ± se (n = 60 guard cells) from three independent experiments.