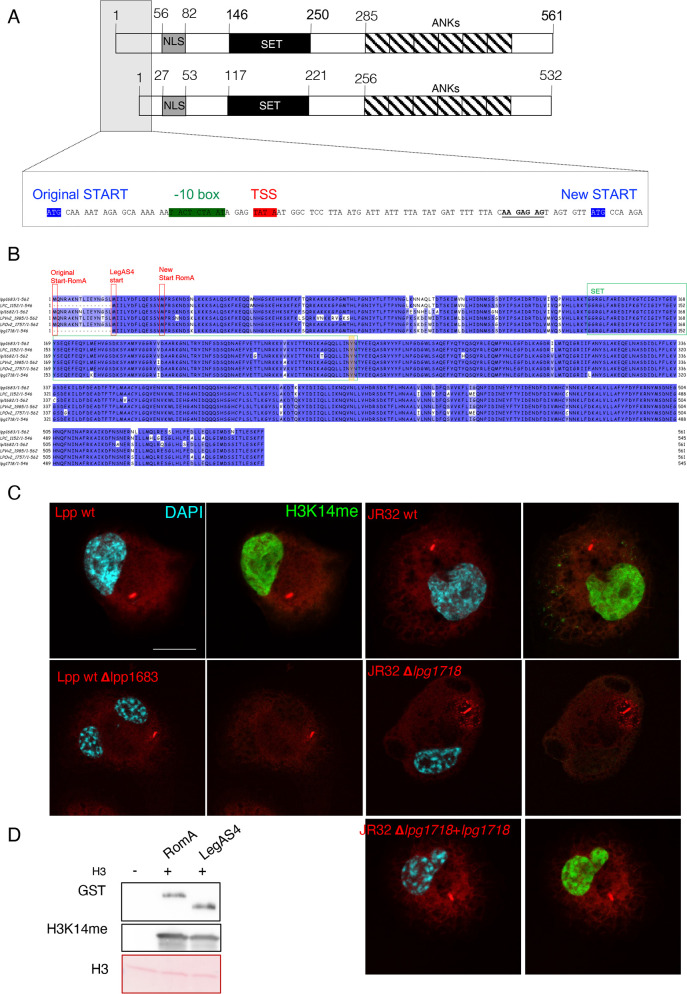
Fig 1.
Sequence and functional comparison of the L. pneumophila effectors RomA and LegAS4. (A) Schematic representation of the RomA protein. NLS, nuclear localization signal; SET, SET domain; ANKs, ankyrin repeats. Insert: the predicted and new start sites identified by TSS mapping are depicted in blue. (B) Protein alignment of RomA homologs in different L. pneumophila strains (lpp, L. pneumophila strain Paris; LPC, L. pneumophila strain Corby; lpl, L. pneumophila strain Lens; LPVv2, L. pneumophila HL06041035; LPOv2, L. pneumophila strain Lorraine; lpg, L. pneumophila strain Philadelphia). Protein starts are highlighted in red; SET domain in green; active site Tyr in orange. (C) Immunofluorescence analysis of THP-1 cells infected 6 hours with wild-type L. pneumophila strain Paris (Lpp wt) and wild-type L. pneumophila strain Philadelphia (JR32 wt) as well with their derivatives, as indicated. JR32-Δlpg1718 strain was complemented with a plasmid encoding the gene lpg1718 under control of its own promoter. Cells were stained for H3K14me2 (green), LPS (red), and DAPI (light blue). Scale bar 10 µm. (D) In vitro methyltransferase activity of RomA or LegAS4 against histone H3; 500 ng of GST-tagged enzymes in the presence of 500 ng of histone H3. The reaction was performed for 30 min at 30°C in the presence of cold SAM. GST-tagged proteins were detected with anti-GST antibodies; methylation of histone H3 by using specific H3K14me antibodies and H3 detected in the ponceau staining.