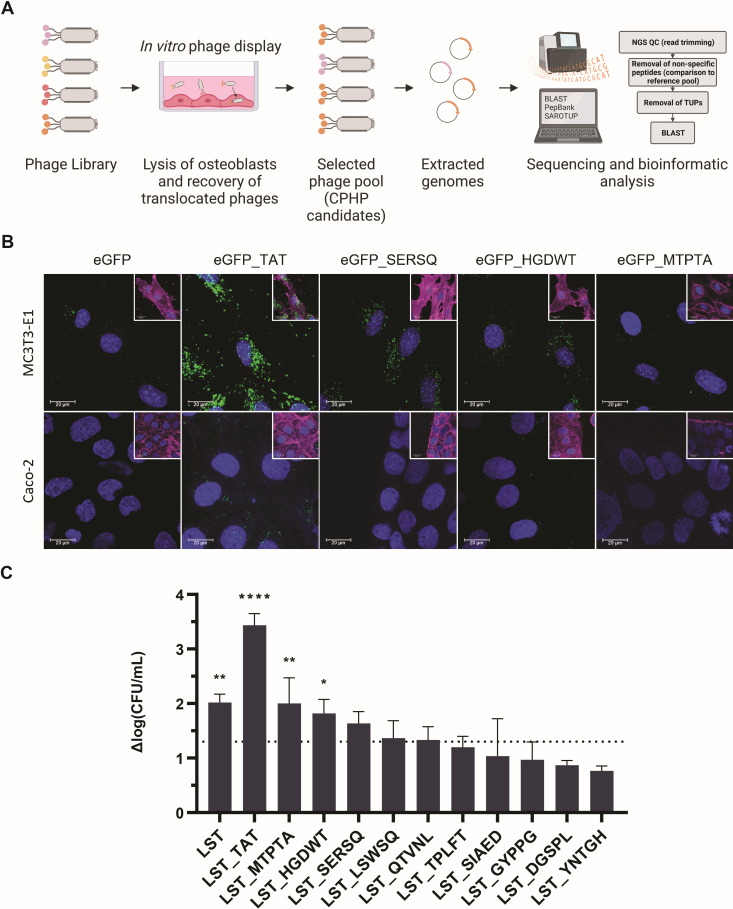
Fig 3.
Selection and characterization of CPHP candidates. (A) CPHP candidates were identified using a cell culture-based phage display approach followed by next-generation sequencing and bioinformatics analysis. Ten CPHP candidates were C-terminally fused to eGFP (B) or LST (C) to assess their cell-line specificity and ability to translocate an active PGH cargo. (B) The cell line-specific uptake of eGFP and CPHP-modified eGFP was assessed by CLSM. MC3T3-E1 (top row) and a non-target cell line (Caco-2, bottom row) were treated for 60 minutes with 5 µM eGFP_CPHP or the controls eGFP and eGFP_TAT (a non-tissue-specific cell-penetrating peptide). Extracellular proteins were removed with three DPBS washes. Cells were stained with FM4-64 (membrane, magenta) and Hoechst 33342 (DNA, blue). Images for the three CPHP candidates that were subsequently used for in vivo experiments are shown. For ease of visualization, an overlay including the stained membrane is shown separately (inset) to the overlay of the eGFP and nucleic acid signals. (C) MC3T3-E1 cells were infected with S. aureus Cowan I (MOI = 5, 1 hour) and treated with LST, LST_TAT, or different LST_CPHP constructs (2 µM, 4 hours). Intracellular bacteria were enumerated by plating, and the average log(CFU/mL) reduction compared to an untreated control was determined (±SEM). The dashed line indicates a 95% reduction in intracellular CFUs. Experiments were conducted in biological triplicates. *P ≤ 0.05; **P ≤ 0.01; and ****P < 0.0001.