Figure 1.
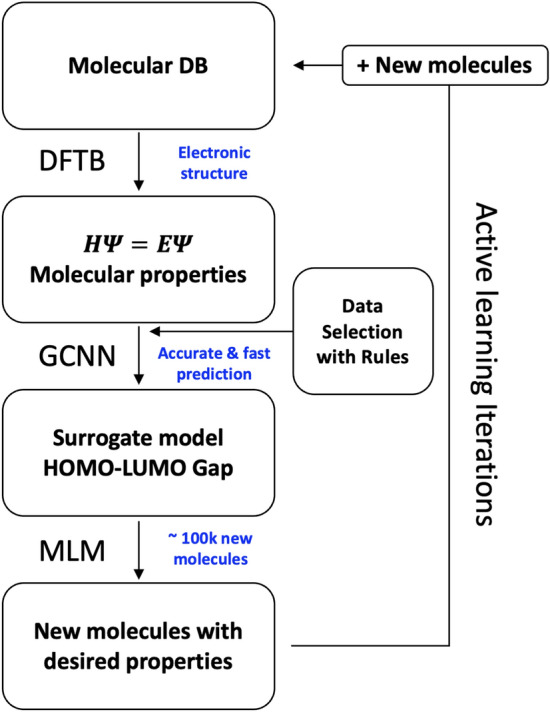
The deep learning iterative workflow for inverse design of molecules with desired properties, namely a specific value of the HLG and high synthesizability. The starting molecular database (DB) in this work was the GDB-9 data set. The DFTB HLGs were taken as ground truth molecular properties. The GCNN surrogate was trained to accurately predict the relationship between molecular structure and HLG. Data selection rules were used to (i) identify newly generated molecules for which the surrogate model shows poor performance for predicting properties, and (ii) to limit the size of the molecule. The MLM mutated and generated new molecules with the trained surrogate model.
