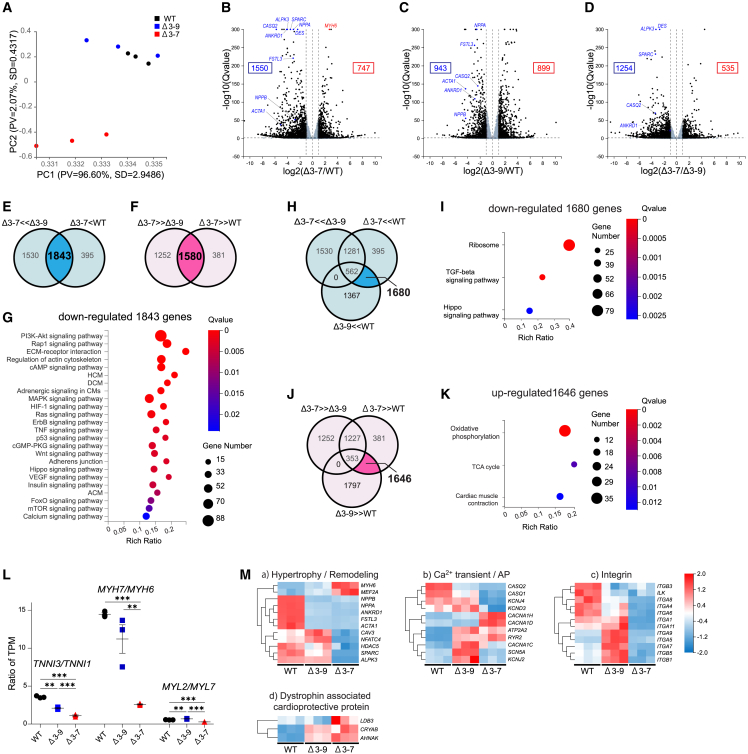
Figure 5.
RNA-seq
(A) PCA of RNA-seq performed on day 48 of WT, Δ3–9, and Δ3–7 hiPSC-CMs (n = 3 independent transductions). The percent variance and SD explained by PC1 (96.60%, SD = 2.9486%) and PC2 (2.07%, SD = 0.4317%) are each listed on the respective axes. (B–D) Volcano plot analysis of Δ3–7 hiPSC-CMs versus WT hiPSC-CMs (B), Δ3–9 hiPSC-CMs versus WT hiPSC-CMs (C), and Δ3–7 hiPSC-CMs versus Δ3–9 hiPSC-CMs (D). The x axis represents the fold change in the difference after conversion to log2, and the y axis represents the significance value after conversion to −log10. Gray represents non-DEGs. (E and F) Venn diagram derived from RNA-seq analysis of hiPSC-CMs from three comparisons between WT, Δ3–9, and Δ3–7 hiPSC-CMs. The threshold of DEGs was adjusted to |Log2FC| ≥1.0 and false discovery rate-adjusted p ≤ 0.05. In Δ3–7 hiPSC-CMs, 1,843 genes were downregulated (E) and 1,580 genes were upregulated (F) compared with those in WT and Δ3–9 hiPSC-CMs. (G) KEGG pathway enrichment analysis of 1,843 downregulated genes in Δ3–7 hiPSC-CMs. (H–K) Venn diagram derived from RNA-seq analysis of hiPSC-CMs from three comparisons between WT, Δ3–9, and Δ3–7 hiPSC-CMs. In both Δ3–7 and Δ3–9 hiPSC-CMs, 1,680 genes were downregulated compared with WT hiPSC-CMs without downregulation in Δ3–7 hiPSC-CMs compared with Δ3–9 hiPSC-CMs. (H) KEGG pathway enrichment of the 1680 genes extracted from (H). (I) In both Δ3–7 and Δ3–9 hiPSC-CMs, 1,646 genes were upregulated compared with WT hiPSC-CMs without upregulation in Δ3–7 hiPSC-CMs compared with Δ3–9 hiPSC-CMs (J). KEGG pathway enrichment of the 1646 genes extracted from (J) (K). (L) FKPM ratio of maturation marker genes, including TNNI3/TNNI1, MYH7/MYH6, and MYL2/MYL7, from WT, Δ3–9, and Δ3–7 hiPSC-CMs. (M) Heatmap of gene normalized z-scores for log2-transformed transcripts per kilobase of exon model per million mapped read values using DEGs involved in hypertrophy and remodeling (a), Ca2+ transient and action potential (AP) (b), integrin (c), and dystrophin-associated cardioprotective protein (d). Data are presented as mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.005.