Fig. 1 |. Single-cell transcriptomics analysis of human breast tissues from BRCA1+/mut and noncarriers.
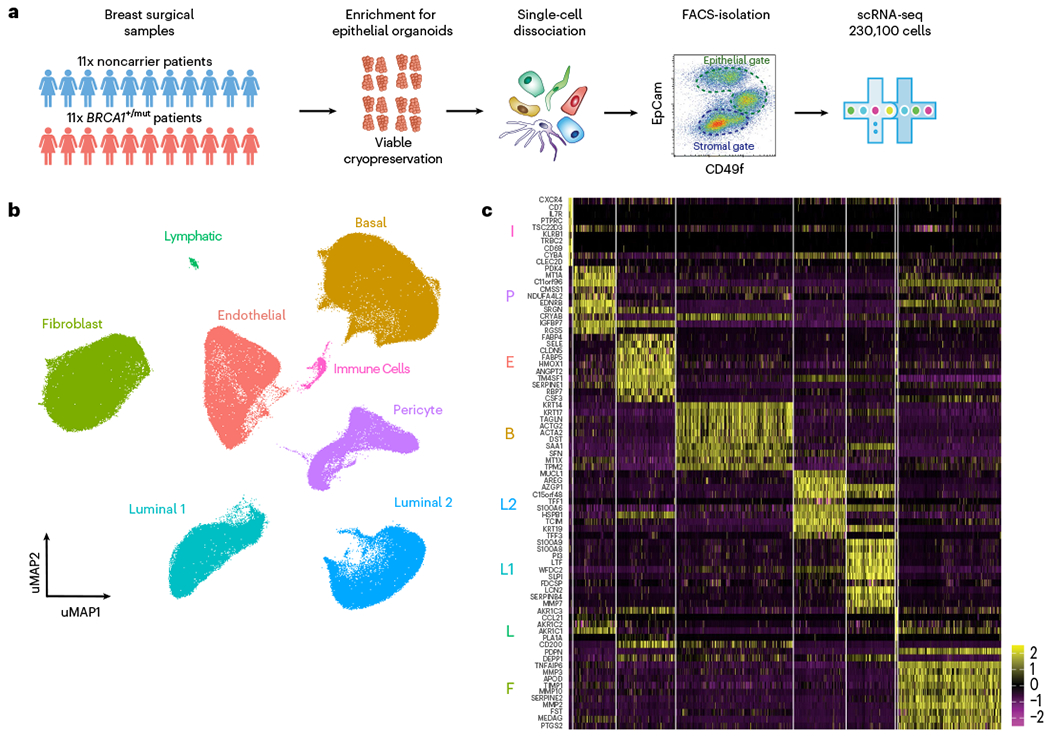
a, Schematic depiction of single-cell analysis workflow using human breast tissue samples that are mechanically and enzymatically dissociated into single-cell suspensions that are subjected to FACS to isolate stromal (EpCAM−) and epithelial (EpCAM+) cells for scRNA-seq analysis. b, Integrated clustering analysis of n = 11 noncarrier and n = 11 BRCA1+/mut scRNA-seq dataset in UMAP projection showing the main identified cell types. c, Top ten marker gene heatmap for each cell type identified by scRNA-seq analysis (rows = genes, columns = cells). The corresponding cell types are indicated with letter abbreviations as follows: basal epithelial cells (B), luminal 1 (L1) and luminal 2 (L2) epithelial cells, fibroblasts (F), pericytes (P), endothelial cells (E), lymphatic cells (L) and immune cells (I).
