Abstract
Iron deficiency (ID) is a widespread condition concomitant with disease and results in systemic dysfunction of target tissues including skeletal muscle. Activated by ID, ferritinophagy is a recently found type of selective autophagy, which plays an important role in various physiological and pathological conditions. In this study, we demonstrated that ID-mediated ferritinophagy impeded myogenic differentiation. Mechanistically, ferritinophagy induced RNF20 degradation through the autophagy-lysosomal pathway and then negatively regulated histone H2B monoubiquitination at lysine-120 in the promoters of the myogenic markers MyoD and MyoG, which inhibited myogenic differentiation and regeneration. Conditional knockout of NCOA4 in satellite cells, overexpression of RNF20 or treatment with 3-methyladenine restored skeletal muscle regenerative potential under ID conditions. In patients with ID, RNF20 and H2Bub1 protein expression is downregulated in skeletal muscle. In conclusion, our study indicated that the ferritinophagy-RNF20-H2Bub1 axis is a pathological molecular mechanism underlying ID-induced skeletal muscle impairment, suggesting potential therapeutic prospects.
Iron deficiency-induced ferritinophagy impairs skeletal muscle regeneration through NCOA4-RNF20-H2Bub1 modification axis.
INTRODUCTION
The trace element iron, which plays a vital role in multiple pathophysiological conditions, is essential for many basic metabolic processes and biochemical activities in cells and organisms (1–4). As a state of iron dyshomeostasis, iron deficiency (ID) is evident in patients with many diseases such as anemia, heart failure, and inflammatory diseases (5–9). Many previous studies have confirmed that iron metabolism is closely related to skeletal muscle differentiation and regeneration (10–13). ID occurs in various chronic diseases, leading to dysfunctional muscle regeneration (10, 12, 14, 15). However, the mechanism by which ID affects muscle differentiation and regeneration is still unknown and needs to be clarified.
Autophagy is a biological recycling process that is highly conserved in eukaryotes. This process involves the removal of unwanted cytoplasmic components and damaged or redundant organelles in eukaryotes under specific physiological or pathological conditions (16–19). Studies have found that under specific pathological conditions, autophagy is selectively activated (20–22). Ferritinophagy is a type of selective autophagy found in recent years. Under ID conditions, ferritinophagy is activated, and ferritin, which is involved in iron storage, is degraded through nuclear receptor coactivator 4 (NCOA4)–mediated ferritinophagy, thus replenishing the concentration of iron ions in cells. Many studies have found that the activation of ferritinophagy under ID conditions contributes greatly to various physiological and pathological conditions (23–27).
Histone H2B monoubiquitination at lysine-120 (H2Bub1) is an important epigenetic modification. H2Bub1 physiologically disrupts the compaction of chromatin, resulting in a more open conformation through which transcription factors and other protein complexes participate in transcriptional activation. This process is extensively involved in the growth and development of the body and the pathological progression of various diseases (28, 29). Histone modification has been reported to modulate nonselective autophagy, and the activation of nonselective autophagy also affects the transcription of downstream genes by regulating the level of histone modification, thereby participating in different physiological and pathological processes (30). However, whether histone H2B monoubiquitination participates in selective autophagy, including ferritinophagy, is still unknown.
The E3 ubiquitin ligase ring finger protein 20 (RNF20) is critical for H2Bub1 activity in mammals (31, 32). Global knockout of RNF20 in mice resulted in profound H2Bub1 loss and caused very early embryonic lethality in vivo (33). Studies have confirmed the essential role of RNF20 in several physiological processes including cell differentiation, spermatogenesis, and adipose tissue development (29, 32, 34). Furthermore, RNF20 deficiency in association with some pathological processes such oncogenesis and inflammation has also been extensively explored (35–37). Although the fundamental role of RNF20 has been recognized under both physiological and pathological conditions, the connection between RNF20 and its downstream target H2Bub1 in the ID context has never been systematically explored.
In this study, we demonstrated that ID triggered ferritinophagy and further inhibited myogenic differentiation by degrading RNF20, thus decreasing the H2Bub1 levels of myogenic markers. Re-establishing RNF20 expression or treating models with 3-methyladenine (3-MA), an autophagy inhibitor, promoted skeletal muscle regeneration under ID conditions in vivo. Conditional knockout of NCOA4 in satellite cells (SCs) showed a protective effect against ID-induced dysfunction of skeletal muscle regeneration. These findings provide insight into the mechanism of ID-induced muscle regeneration impairment.
RESULTS
ID-mediated ferritinophagy inhibits myoblast differentiation
We first induced ID in human muscle primary SCs with the classic iron-chelating agent deferoxamine (DFO). Western blot analysis showed that as the DFO concentration increased, the protein levels of NCOA4 and ferritin heavy chain (FTH) gradually decreased, while the LC3II levels increased (Fig. 1A). The confocal immunofluorescence results were similar to those of the Western blot analysis. As the DFO concentration increased, the fluorescence intensity of NCOA4 gradually decreased, while LC3B accumulation was enhanced (Fig. 1B). These results indicated that DFO activates ferritinophagy in human muscle primary SCs, which is consistent with previous reports (23). Furthermore, as the DFO concentration increased, the expression levels of myogenic markers, including MyHC, MyoD, and MyoG, decreased in a dose-dependent manner at both the mRNA and protein levels (Fig. 1, C and D). Immunofluorescence analysis showed that with increasing DFO concentration, the fusion index and average myotube length was decreased, while the fluorescence intensity of MyoD and MyoG, key transcription factors in muscle differentiation, was weaker (Fig. 1E). These results suggested that ID inhibits the myogenic differentiation of muscle SCs. Then, we added ammonium ferric citrate (AFC) to restore intracellular iron or the classic autophagy inhibitors DC661 and 3-MA to determine whether blocking ferritinophagy restores myoblast differentiation. Western blot analysis showed that treatment with AFC, DC661, and 3-MA reversed the DFO-mediated decrease in myogenic marker expression (Fig. 1F), and confocal immunofluorescence showed that treatment with AFC, DC661, and 3-MA partially restored the fusion index and average myotube length (Fig. 1G). Moreover, we down-regulated NCOA4 or ATG7 expression using small interfering RNAs (siRNAs) to block ferritinophagy activity. Western blot analysis showed that NCOA4 and ATG7 were effectively knocked down and that DFO-mediated myogenic differentiation inhibition was abrogated (Fig. 1H). Confocal immunofluorescence of MyHC also showed that down-regulating NCOA4 or ATG7 expression effectively restored the level of muscle fiber formation (Fig. 1I). We repeated the abovementioned experiment with C2C12 cells (fig. S1). Besides, no significant difference in the viability of C2C12 cells treated with DFO at concentrations from 10 to 50 μM was found (fig. S2A). However, DFO exerted a slight proliferation-inhibiting effect at high concentrations (fig. S2B). In the absence of DFO, AFC, DC661, and 3-MA exerted slight inhibitory effects on MyHC, MyoD, and MyoG expression in C2C12 cells (fig. S3, A and B). Moreover, similar results were obtained in C2C12 cells treated with another iron-chelating agent, deferiprone (DFP) (fig. S4). Together, these results suggested that ID inhibits myoblast differentiation by triggering ferritinophagy.
Fig. 1. Iron deficiency activates ferritinophagy and inhibits myogenic differentiation in human muscle primary SCs.
(A) Western blot analysis of NCOA4, FTH, and LC3B expression in primary SCs treated with 0, 10, 15, 20, 25, and 30 μM DFO in differentiation medium for 3 days (n = 9). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Immunofluorescence staining for NCOA4 (green) and LC3B (red) in DFO-treated or untreated primary SCs cultured for 3 days in differentiation medium (n = 9). Scale bars, 50 μm. DAPI, 4′,6-diamidino-2-phenylindole; MFI, mean fluorescence intensity. (C) mRNA levels of MyHC, MyoD, and MyoG in primary SCs treated with 0, 10, 15, 20, 25, and 30 μM DFO in differentiation medium for 3 days (n = 9). (D) Western blot analysis of MyHC, MyoD, and MyoG expression in primary SCs treated with 0, 10, 15, 20, 25, and 30 μM DFO in differentiation medium for 3 days (n = 9). (E) Immunofluorescence staining for MyHC, MyoD, and MyoG in primary SCs treated with 0, 10, 20, and 30 μM DFO in differentiation medium for 3 days (n = 9). Scale bars, 100 μm. (F) Western blot analysis of MyHC, MyoD, and MyoG expression in primary SCs treated as indicated in differentiation medium for 3 days (n = 9). (G) Immunofluorescence staining for MyHC in primary SCs treated as indicated in differentiation medium for 3 days (n = 9). Scale bars, 100 μm. (H) Western blot analysis of ATG7, NCOA4, MyHC, MyoD, and MyoG expression in primary SCs treated as indicated in differentiation medium for 3 days (n = 9). n.s., not significant. (I) Immunofluorescence staining for MyHC in primary SCs treated as indicated in differentiation medium for 3 days (n = 9). Scale bars, 100 μm. The results represent the means ± SD. *P < 0.05 and **P < 0.01.
Ferritinophagy degrades RNF20 and negatively regulates H2Bub1 modification
To further explore the mechanism by which ferritinophagy regulates myoblast differentiation, we first explored the proteins binding to the ferritinophagy-specific cargo receptor NCOA4 after activation of ferritinophagy through coimmunoprecipitation (Co-IP) and liquid chromatography tandem mass spectrometry (LC-MS/MS). The results showed that 185 proteins were bound to NCOA4 in C2C12 cells after the activation of ferritinophagy (Supplementary Materials). Notably, RNF20 was involved in the Co-IP complex (Fig. 2A). RNF20 is an E3 ubiquitin ligase that specifically mediates H2Bub1 modification (38, 39). We then conducted reciprocal Co-IP/Western blot assays to confirm that endogenous NCOA4 and RNF20 interacted with each other in C2C12 cells (Fig. 2B) and that exogenous Myc-NCOA4 and Flag-RNF20 interacted with each other in 293T cells (fig. S5A). Moreover, immunofluorescence assays showed that NCOA4 colocalized with RNF20 in lysosomes after the activation of ferritinophagy (fig. S6). In addition, RNF20 did not interact with other autophagy-related cargo receptor molecules after the activation of ferritinophagy (fig. S5B). These results indicated that the binding of NCOA4 to RNF20 is relatively specific.
Fig. 2. Ferritinophagy degrades RNF20 and negatively regulates H2Bub1 modification.
(A) NCOA4 interacts with RNF20 as determined by Coomassie bright blue staining. IgG, immunoglobulin G; IP, immunoprecipitation; WCL, whole-cell lysate. (B) Interaction between endogenous NCOA4 and RNF20 in C2C12 cells was confirmed by Co-IP with Western blotting (n = 9). (C) Western blot analysis of RNF20 and H2Bub1 expression in C2C12 cells treated with 0, 10, 15, 20, 25, and 30 μM DFO in differentiation medium for 3 days (n = 9). (D) Western blot analysis of RNF20 and H2Bub1 in C2C12 cells treated as indicated in differentiation medium for 3 days (n = 9). (E) Western blot analysis of NCOA4, RNF20, and H2Bub1 in C2C12 cells treated as indicated in differentiation medium for 3 days (n = 9). The results represent the means ± SD. *P < 0.05 and **P < 0.01.
Next, we explored whether the expression levels of RNF20 and H2Bub1 change after the activation or inhibition of ferritinophagy. Western blot analysis demonstrated that the protein levels of RNF20 and H2Bub1 were decreased in a dose-dependent manner with DFO treatment (Fig. 2C). However, quantitative polymerase chain reaction (qPCR) analysis showed that there was no change in RNF20 gene expression after DFO treatment (fig. S5C). Therefore, we hypothesized that after the activation of ferritinophagy, RNF20 may be degraded through the autophagy-lysosomal pathway, thereby regulating the level of H2Bub1. Western blot analysis showed that the addition of autophagy inhibitors inhibited DFO-mediated down-regulation of RNF20 and H2Bub1 expression, while the addition of the proteasome inhibitor MG132 did not have a similar effect (Fig. 2D). Furthermore, we detected the regulatory effect of ferritinophagy on RNF20 expression using NCOA4 or ATG7 siRNA. Western blot results showed that NCOA4 and ATG7 expression was decreased, and DFO-mediated down-regulation of RNF20 and H2Bub1 levels was reversed (Fig. 2E). Last, we examined the changes in the expression levels of other H2B monoubiquitination-specific ubiquitination ligases and deubiquitination enzymes during the activation of ferritinophagy. Western blot analysis showed that the expression levels of the other enzymes did not change after the activation of ferritinophagy (fig. S5, D and E). These results indicated that the activation of ferritinophagy leads to RNF20 degradation mediated through the autophagy-lysosomal pathway and decreases the level of H2Bub1.
RNF20 positively regulates muscle formation by modulating H2Bub1 modification of MyoD and MyoG
To determine the role of the RNF20 and H2Bub1 axes in myoblast differentiation, we designed an siRNA to knock down endogenous RNF20 expression. qPCR and Western blot analyses demonstrated that the expression of MyHC, MyoG, and MyoD was decreased at both the mRNA and protein levels after RNF20 was knocked down. H2Bub1 and myogenesis marker levels were also decreased after RNF20 siRNA treatment (Fig. 3, A and B). In addition, immunofluorescence assays demonstrated that the fusion index and average myotube length was decreased after RNF20 was knocked down (Fig. 3C). These results demonstrated that RNF20 positively regulates myogenic differentiation.
Fig. 3. RNF20 positively regulates muscle formation by modulating H2Bub1 modification of MyoD and MyoG.
(A) mRNA expression of RNF20, MyHC, MyoD, and MyoG was measured by qRT-PCR after knockdown of RNF20 by siRNA in C2C12 cells cultured for 3 days in differentiation medium (n = 9). (B) Western blot analysis of RNF20, H2Bub1, MyHC, MyoD, and MyoG protein expression in the RNF20 knockdown or control group of C2C12 cells cultured for 3 days in differentiation medium (n = 9). (C) Immunofluorescence staining for MyHC in the RNF20 knockdown and control group of C2C12 cells cultured for 3 days in differentiation medium (n = 9). Scale bars, 100 μm. (D) mRNA expression of RNF20, MyHC, MyoD, and MyoG in the RNF20 overexpression group of C2C12 cells treated with or without DFO (20 μM) (n = 9). (E) Western blot analysis of RNF20, H2Bub1, MyHC, MyoD and MyoG expression in the RNF20 overexpression group of C2C12 cells treated with or without DFO (20 μM) (n = 9). (F) Immunofluorescence staining for MyHC in the RNF20 overexpression group of C2C12 cells treated with or without DFO (20 μM) (n = 9). Scale bars, 100 μm. (G) Results of ChIP-qPCR confirmed changes in H2Bub1 modification and binding to the promoter of MyoD and MyoG in control and iron-deficient C2C12 cells treated with or without DC661 following the induction of differentiation for 3 days (n = 9). (H) ChIP-qPCR analysis of changes in H2Bub1 binding to the promoters of MyoD and MyoG in RNF20-overexpressing C2C12 cells under DFO (20 μM), treated or untreated (n = 9). The results represent the means ± SD. *P < 0.05 and **P < 0.01. ID, iron-deficient.
Furthermore, we constructed an RNF20 overexpression lentivirus. qPCR and Western blot analyses demonstrated that overexpression of RNF20 reversed the expression of MyHC, MyoG, and MyoD at both the mRNA and protein levels as well as the H2Bub1 level under ID conditions (Fig. 3, D and E). Moreover, RNF20 overexpression reversed the impaired muscle fiber formation by C2C12 cells under DFO-induced ID conditions (Fig. 3F).
A previous study demonstrated that RNF20 regulates target gene expression by modulating H2Bub1. We further performed chromatin immunoprecipitation (ChIP)–qPCR analysis to explore RNF20 binding and H2Bub1 signals at the promoters of the MyoG and MyoD myogenic transcription factors. Compared to those of the control group, the binding signals of RNF20 and H2Bub1 at the promoters of MyoG and MyoD were reduced under ID conditions. Furthermore, blocking ferritinophagy with DC661 reversed the binding signal intensity at the promoters of MyoG and MyoD under ID conditions (Fig. 3G). Moreover, overexpression of RNF20 reversed the binding signaling intensity (Fig. 3H). In conclusion, these results indicated that RNF20 increases the H2Bub1 level at MyoG and MyoD promoters, positively regulating MyoG and MyoD gene expression and subsequent muscle formation.
We further recruited six individuals with ID (serum iron of <50 μg/dl) and six age- and sex-matched healthy controls with normal iron levels (serum iron of >50 μg/dl) (table S1). The hemoglobin, hematocrit, serum iron, and transferrin saturation levels were measured (fig. S7, A and B). The iron level in muscles of patients with ID was lower than that in the control group individuals (fig. S7C). Levels of the iron storage protein ferritin and the ferritinophagy-specific cargo receptor NCOA4 were decreased in the skeletal muscle of the iron-deficient patient group (fig. S7D). Moreover, RNF20 and H2Bub1 expression was down-regulated in the skeletal muscle of the iron-deficient patient group compared to the healthy control group (fig. S7, E and F).
RNF20 and 3-MA protect against ID-induced delay in skeletal muscle growth
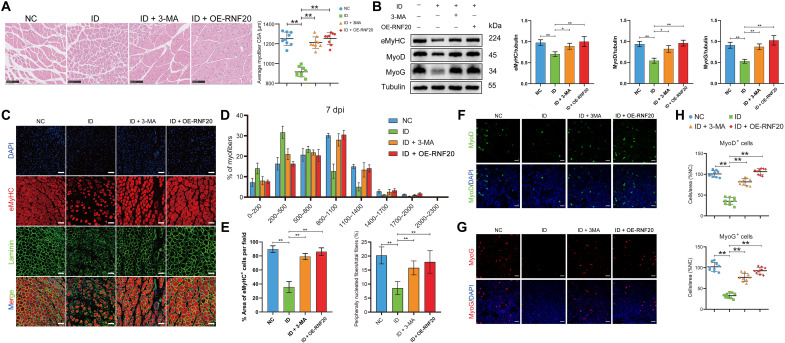
To investigate the effect of blocking ferritinophagy or increasing the expression of RNF20 on ID-induced delay in skeletal muscle growth in vivo, we constructed an iron-deficient mouse model. Mice aged 3 to 5 weeks were fed an iron-deficient diet for 8 weeks. Compared with that of the normal group mice, the body weight of the mice fed an iron-deficient diet was decreased (fig. S8A). The hemoglobin, hematocrit, serum iron, and transferrin saturation levels indicated that the experimental mice exhibited ID, in contrast to the control mice (fig. S8, B to E). As determined by laser ablation–inductively coupled plasma–mass spectrometry (LA-ICP-MS), iron-deficient mice had less iron in their skeletal muscle compared to the amount in the control mice (Fig. 4A), and this effect was confirmed via colorimetric assay (fig. S8F). Decrease in the ratio of gastrocnemius (GA) muscle weight to body weight was observed in the iron- deficient group compared to the control group (Fig. 4B and fig. S8G). Moreover, intraperitoneal administration of 3-MA or intramuscular injection of RNF20-overexpressing adeno-associated virus (AAV) in mice reversed the changes in muscle volume and the ratio of GA muscle weight to body weight (Fig. 4B and fig. S8G). Functionally, iron-deficient mice exhibited decreased forelimb grip strength, inverted hanging time, and treadmill exhaustive running time than those in the control group. Meanwhile, intraperitoneal administration of 3-MA or intramuscular injection of RNF20-overexpressing AAV into the mice could reverse the decline in the skeletal muscle function in iron-deficient mice (Fig. 4C). GA muscle sections were further collected for histological analysis to assess the effect of ID on skeletal muscle in mice. Iron levels were assessed by Prussian blue staining, which indicates the non-heme iron level in tissues. The iron level was lower in iron- deficient mice compared to control mice (Fig. 4D). Hematoxylin and eosin (H&E) staining showed that the cross-sectional area (CSA) of the GA muscle fibers in the iron-deficient group was smaller than that in the normal diet group (Fig. 4D). After the intraperitoneal administration of 3-MA or intramuscular injection of RNF20-overexpressing AAV, the CSA of muscle was improved (Fig. 4D). Western blot and immunofluorescence analyses showed that the expression of the MyoG and MyoD myogenic markers was inhibited in the iron-deficient group, while this change was reversed in the 3-MA– or AAV-overexpressing RNF20 group (Fig. 4, E to H). Besides, in the absence of ID, 3-MA treatment slightly decreased the muscle size, and overexpression of RNF20 exerted no effect on muscle size. Similar results were observed in the MyoD and MyoG immunofluorescence assays (fig. S3, C to E).
Fig. 4. RNF20 and 3-MA protect against iron deficiency–induced delay in skeletal muscle growth.
Mice were divided into the following four groups with nine animals per group: normal control group (NC), iron-deficient group, ID plus 3-MA (ID + 3-MA) group, and ID plus RNF20 overexpression (ID + OE-RNF20) group. (A) Iron levels in the skeletal muscle tissue were measured via LA-ICP-MS and in situ imaging analysis. (B) Representative images of GA muscles in NC mice, iron-deficient mice, ID + 3-MA mice, and ID + OE-RNF20 mice. (C) Skeletal function was estimated by tests of grip strength, hanging time, and exhaustion time (n = 9). (D) H&E staining and Prussian blue staining for iron in the GA muscle of different groups of mice (n = 9). Scale bars in H&E staining, 100 μm. Scale bars in Prussian blue staining, 50 μm. (E) Western blot analysis of MyoD and MyoG expression in GA muscle samples from different groups of mice (n = 9). (F to H) Immunofluorescence analysis of MyoD and MyoG expression in the GA muscle of different groups of mice (n = 9). Scale bar, 50 μm. The results represent the means ± SD. *P < 0.05 and **P < 0.01. ID, iron-deficient.
The expression of RNF20 and H2Bub1 in vivo was verified. Western blot and immunofluorescence analyses demonstrated that the expression of RNF20 and H2Bub1 in muscle was decreased in the iron-deficient group, and this change was reversed by 3-MA or AAV overexpression of RNF20 (Fig. 5, A to C). In summary, these results indicated that ID-induced skeletal muscle growth delay mediated via ferritinophagy leads to a decrease in RNF20-H2Bub1 levels in vivo.
Fig. 5. RNF20 and H2Bub1 levels in iron-deficient mice after 3-MA treatment and RNF20 overexpression.
(A and B) Immunofluorescence staining for RNF20 and H2Bub1 in different groups of mice. The mean fluorescence intensities of RNF20 and H2Bub1 are shown below (n = 9). Scale bars, 50 μm. (C) Western blot analysis of RNF20 and H2Bub1 expression in different groups of mice (n = 9). The results represent the means ± SD. **P < 0.01. ID, iron-deficient.
RNF20 and 3-MA promote skeletal muscle regeneration under ID conditions
To further verify the role of ID in the skeletal muscle regeneration, we used a typical cardiotoxin (CTX) in GA muscles to perform skeletal muscle regeneration experiments in iron-deficient mice. H&E staining analysis showed that after 7 days of muscle regeneration, the control group had formed many muscle fibers, while the skeletal muscle of the iron-deficient mice regenerated fewer muscle fibers (Fig. 6A). Overexpression of RNF20 and the addition of 3-MA promoted skeletal muscle regeneration under ID conditions (Fig. 6A). The expression of eMyHC, MyoD, and MyoG was lower in the iron-deficient group than in the control group, but the expression was rescued in the 3-MA and RNF20 overexpression groups (Fig. 6B). Immunostaining analysis showed that the proportion of eMyHC+ fibers, which represent newly regenerated muscle fibers, was lower in the iron-deficient group than in the control group. In addition, the GA muscle of the iron-deficient mice had a smaller fiber distribution compared with that of the control group, as demonstrated by laminin staining and determination of the fiber CSA. Treatment with 3-MA or overexpression of RNF20 reversed the formation of eMyHC+ fibers and the peripheral fiber nucleation rate (Fig. 6, C to E). The immunostaining of MyoG+ cells and MyoD+ cells showed similar results (Fig. 6, F to H). In absence of ID, 3-MA treatment led to a slight decrease in muscle size and MyoD and MyoG immunofluorescence intensity, and the overexpression of RNF20 exerted no effect (fig. S3, F to H).
Fig. 6. RNF20 and 3-MA promote skeletal muscle regeneration under iron deficiency conditions.
The mice were divided into the following four groups with nine mice per group: NC, iron-deficient group, ID + 3-MA group, and ID + OE-RNF20 group. The CTX was injected into GA muscles of the above mice to induce acute injury. The injected GA muscles were harvested at the 7 day post-CTX injury (dpi) for the assessment of the regeneration process. (A) H&E staining of the GA muscle and the average myofiber CSA in different groups of mice at 7 dpi (n = 9). Scale bars, 100 μm. (B) Western blot analysis of eMyHC, MyoD, and MyoG expression in different groups of mice at 7 dpi (n = 9). (C) Immunofluorescence analysis of eMyHC+ fibers in the GA muscle of different groups of mice at 7 dpi. Scale bars, 50 μm. (D) Percent distribution of the muscle fiber cross-sectional area derived from the different groups of mice at 7 dpi (n = 9). (E) eMyHC+ cells and peripherally nucleated fibers in the GA muscle of different groups of mice at 7 dpi (n = 9). (F to H) Immunofluorescence analysis of MyoD+ and MyoG+ cells in the GA muscle of different groups of mice at 7 dpi (n = 9). Scale bars, 20 μm. The results represent the means ± SD. *P < 0.05 and **P < 0.01. ID, iron-deficient.
Western blot and immunofluorescence analyses demonstrated that under ID conditions, the RNF20 and H2Bub1 levels were decreased, and this trend was reversed after adding 3-MA or overexpressing RNF20 (fig. S9, A to C). In conclusion, these results indicated that ID inhibits skeletal muscle regeneration, which is reversed by 3-MA and RNF20 in vivo.
Genetic deletion of NCOA4 in SCs restores ID-induced skeletal muscle regeneration dysfunction
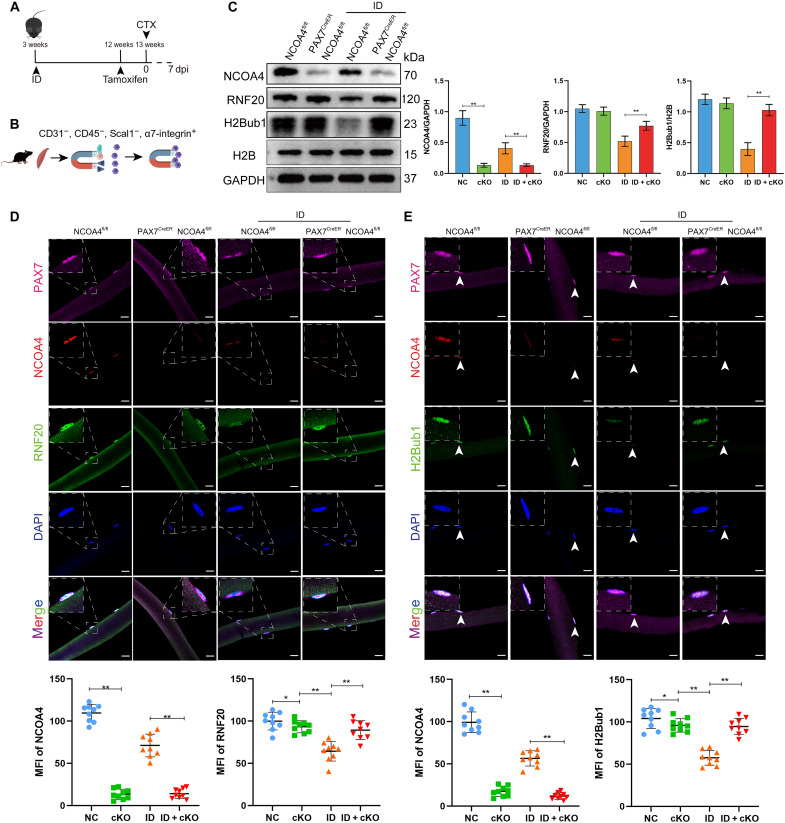
SC-specific NCOA4 knockout mice were generated by crossing mice having the NCOA4fl/fl conditional allele with mice expressing a CreER protein in Pax7+ cells. The Pax7-CreER, NCOA4fl/fl and NCOA4fl/fl genotypes were used. NCOA4 conditional knockout (cKO) mice and control littermates were named NCOA4SCs/KO and NCOA4SCs/WT, respectively. The iron-deficient mouse model was constructed using NCOA4SCs/KO and NCOA4SCs/WT as described above. NCOA4 deletion was induced by the intraperitoneal injection of tamoxifen (TMX) for seven consecutive days (Fig. 7A). To evaluate the recombination efficiency at the protein level, we analyzed magnetic-activated cell sorting (MACS)–purified SCs that were CD31−, CD45−, Scal−, and α7-integrin+ from NCOA4SCs/KO and NCOA4SCs/WT mice (Fig. 7B) and found that NCOA4 expression was inhibited in NCOA4SCs/KO mice. Meanwhile, the expression of RNF20 and H2Bub1 was decreased in the iron-deficient group of NCOA4SCs/WT mice, and this change was reversed after NCOA4 was conditionally knocked out (Fig. 7C). Furthermore, the fluorescence intensities of RNF20 and H2Bub1 were reduced in Pax7+ cells under ID conditions, and knocking out NCOA4 offset the decline in RNF20 and H2Bub1 (Fig. 7, D and E).
Fig. 7. Genetic deletion of NCOA4 in SCs restores the expression of RNF20 and H2Bub1.
The mice were divided into the following four groups with nine mice per group: NC group (NCOA4fl/fl), cKO group (Pax7CreER, NCOA4fl/fl), iron-deficient group (NCOA4fl/fl) group, and ID + cKO group (Pax7CreER, NCOA4fl/fl) group. (A) Schematic diagram of the procedures for iron deficiency induction, NCOA4 cKO, and CTX-induced muscle injury. (B) Schematic diagram of SC isolation. (C) Western blot analysis of NCOA4, RNF20, and H2Bub1 protein levels in GA muscle samples from different groups of mice at 7 dpi (n = 9). (D) Representative images of myofibers isolated from the extensor digitorum longus (EDL) muscle of NCOA4SCs/WT and NCOA4SCs/KO mice (n = 9). Immunofluorescence staining of Pax7 (pink), NCOA4 (red), RNF20 (green), and DAPI (blue). Scale bars, 20 μm. (E) Representative images of myofibers isolated from the EDL muscle of NCOA4SCs/WT and NCOA4SCs/KO mice (n = 9). Immunofluorescence of Pax7 (pink), NCOA4 (red), H2Bub1 (green), and DAPI (blue) staining. Scale bars, 20 μm. The results represent the means ± SD. *P < 0.05 and **P < 0.01. ID, iron-deficient.
A muscle injury model with ID was also constructed using NCOA4SCs/KO and NCOA4SCs/WT mice. H&E staining analysis showed that after 7 days of CTX injection, the skeletal muscle of the iron-deficient group regenerated fewer muscle fibers than the control group. Knocking out NCOA4 in SCs promoted skeletal muscle regeneration under ID conditions (Fig. 8A). The expression of eMyHC, MyoD, and MyoG was lower in the iron-deficient group than in the control group, but the expression of these markers was rescued in the NCOA4SCs/KO groups (Fig. 8B). Immunostaining analysis showed that the proportion of eMyHC+ fibers, which represent newly regenerated muscle fibers, was lower in the iron-deficient group than in the control group. Knocking out NCOA4 reversed the formation of eMyHC+ fibers (Fig. 8C). Compared with that of the control group, the GA muscle of iron-deficient mice showed a smaller fiber distribution, as demonstrated by laminin staining and measurements of the CSA. In NCOA4SCs/KO mice, the larger fibers and the eMyHC+ fibers as well as the peripheral fiber nucleation rate were represented, similar to control mice (Fig. 8, D and E). Consistently, immunostaining of MyoG+ cells and MyoD+ cells showed similar results (Fig. 8, F to H).
Fig. 8. Genetic deletion of NCOA4 in SCs reverses skeletal muscle regeneration defects.
(A) H&E staining of the GA muscle in different groups of mice at 7 dpi (n = 9). Scale bars, 100 μm. (B) Western blot analysis of eMyHC, MyoD, and MyoG protein levels in GA muscle samples from different groups of mice at 7 dpi (n = 9). (C) Immunofluorescence analysis of eMyHC+ fibers in the GA muscle of different groups of mice at 7 dpi. Scale bars, 50 μm. (D) Percent distribution of the muscle fiber cross-section area derived from NCOA4SCs/WT and NCOA4SCs/KO mice at 7 dpi (n = 9). (E) The eMyHC+ cells and peripherally-nucleated fibers in the GA muscle of different groups of mice at 7 dpi (n = 9). (F and G) Immunofluorescence analysis of MyoD+ and MyoG+ cells in the GA muscle of different groups of mice at 7 dpi. Scale bars, 20 μm. (H) The percentage of MyoD+ and MyoG+ cells at 7 dpi (n = 9). The results represent the means ± SD. *P < 0.05 and **P < 0.01. ID, iron-deficient.
DISCUSSION
The present study explored how ID-activated ferritinophagy affects skeletal muscle differentiation and regeneration, and we found that RNF20-mediated histone H2B monoubiquitination plays an important role in this process. When ID activates ferritinophagy in SCs and C2C12 cells, the ferritinophagy cargo receptor NCOA4 interacts with the histone E3 ubiquitin ligase RNF20, promoting RNF20 degradation through the autophagy-lysosomal pathway, which reduces the histone H2B monoubiquitination level at the promoters of the MyoD and MyoG myogenic transcription factors and subsequently leads to inhibition of myogenic differentiation. Overexpressing RNF20 and adding the autophagy inhibitor 3-MA reversed the skeletal muscle regeneration dysfunction in mice with ID in vivo. Pax7-CreER, NCOA4fl/fl mice were constructed, and the cKO of NCOA4 in SCs was shown to reverse ID-induced skeletal muscle regeneration defects. Moreover, NCOA4 and RNF20 expression down-regulation and H2Bub1 level decrease were confirmed in patients with ID.
Iron is one of the most important ions in the human body. An adequate amount of iron is essential for body homeostasis, while either iron overload or deficiency establishes pathological conditions in vivo. In recent years, extensive research has been conducted to determine the contribution of iron accumulation on myogenic pathology, such as muscle regeneration dysfunction mediated through oxidative stress and lipid peroxidation (11, 13, 40). In addition to iron overload, ID can also lead to skeletal muscle regeneration impairment (10, 12, 15, 41, 42). However, in contrast to that underlying iron overload, the mechanism by which ID affects muscle regeneration has not been fully elucidated. Ferritinophagy is a type of selective autophagy. When the iron supply in the body is sufficient, iron is stored in ferritin in cells to reduce the labile iron pool. In contrast, under conditions of ID, ferritin is degraded by selective autophagy, releasing iron ions into the cell (26, 27). Under physiological conditions, ferritinophagy dynamically regulates intracellular iron levels by regulating ferritin expression to maintain a steady state of iron ions in the cell. However, under pathological conditions, the ID status in the body is maladjusted, and ferritinophagy is abnormally activated. For example, the activation of ferritinophagy promotes ferroptosis and plays an important role in progression of neurodegenerative diseases, such as Parkinson’s disease and Alzheimer’s disease (25, 43). Bauckman and colleagues also reported that ferritinophagy increases susceptibility to infectious diseases (44, 45). In our research, we demonstrated that ID activated ferritinophagy, thereby inhibiting the differentiation of myoblasts and regeneration of skeletal muscle, and these effects were reversed by inhibiting ferritinophagy. Therefore, our study suggested that activation of ferritinophagy may be an important factor in skeletal muscle regeneration dysfunction in iron-deficient patients with any of a variety of chronic diseases. These findings clarify the mechanism underlying muscle regeneration dysfunction in patients with ID.
Autophagy is a biological recycling process that is highly conserved in eukaryotes. This process involves the removal of unwanted cytoplasmic components and damaged and/or redundant organelles in eukaryotes under specific physiological or pathological conditions (16–19). Previously, the activation of autophagy was believed to be nonselective. When autophagy is activated, many different types of cell components are degraded by the autophagy-lysosomal pathway. However, recent studies have found that under certain pathological conditions, the activation of autophagy—including mitophagy, xenophagy, reticulophagy and lipophagy—is often selective and leads to the relatively specific degradation of cargo proteins through specific receptor proteins (20, 21, 22, 44, 46). NCOA4 is a cargo receptor in ferritinophagy, and it interacts with ferritin, promoting its transport to autophagosomes, where it is degraded. Under ID conditions, ferritinophagy is activated, causing NCOA4 to bind to and promote the degradation of iron-binding proteins through the autophagic pathway, which replenishes the concentration of iron ions in cells (23, 24, 26, 27). In our study, we demonstrated that during activation of ferritinophagy, NCOA4 not only bound FTH for degradation but also bound RNF20 for degradation, further modulating the level of H2Bub1. Moreover, this process is relatively selective, as RNF20 bound to NCOA4 but did not bind to other autophagy cargo receptors, such as NBR1 or Optineurin.
Some studies have explored the effect of autophagy on muscle differentiation and regeneration. However, the results are controversial, as some studies have demonstrated that autophagy promotes myogenic differentiation and regeneration, while others have indicated that autophagy negatively controls myogenic differentiation and regeneration (47–51). We believe that these contradictory results are due to the outdated concept that autophagy is nonselective. In these various studies, the conditions for autophagic activation were completely different. Thus, the downstream cargo and cargo receptors were different, resulting in different biological effects. Therefore, we suggest that future studies on autophagy should be specifically focused on the activation of selective autophagy under different conditions.
Here, we demonstrated that ferritinophagy inhibited muscle differentiation mainly through lysosome-mediated RNF20 degradation. RNF20 is a ring finger–type E3 ubiquitin ligase, and the structure of RNF20 has been highly conserved during evolution. RNF20 functions mainly through H2Bub1 in mammalian cells to promote RNA polymerase II–mediated mRNA transcription to activate gene transcription and expression (26, 38, 39). Previous studies have shown that RNF20 plays an important role in the differentiation of multiple mesenchymal cells, including osteogenic differentiation, adipogenic differentiation, and astrocytic differentiation (29, 32, 52, 53). Moreover, the present study demonstrated that RNF20 positively regulated myogenic differentiation. ChIP-qPCR further showed that RNF20 knockdown led to decreased binding intensity of H2Bub1 at the promoter of the essential myogenic transcription factors MyoG and MyoD, which decreased MyoG and MyoD transcription and thus inhibited myogenic differentiation. Moreover, RNF20-overexpressing AAV not only protected against ID-induced skeletal muscle growth delay but also promoted skeletal muscle regeneration under ID conditions. Thus, our study further supports a role for RNF20 in mesenchymal cell differentiation. A previous study showed that starvation-induced autophagy mainly affects the formation of H2B monoubiquitination by up-regulating ubiquitin-specific protease 44 (USP44) expression (30). However, we found that during the activation of ferritinophagy, USP44 expression remained stable, while RNF20 was degraded by the NCOA4-mediated autophagy-lysosomal pathway, resulting in a decrease in the H2B monoubiquitination level. This interesting phenomenon further demonstrated that the activation of autophagy under different conditions is relatively selective.
We further demonstrated the role of the ID-induced ferritinophagy- related RNF20-H2Bub1 axis in skeletal muscle regeneration in vivo. The results showed that ID decreased the expression of MyoG and MyoD in muscle and impaired muscle regeneration in vivo. To further verify that ID impaired skeletal muscle regeneration through ferritinophagy degrading RNF20, we added 3-MA to block ferritinophagy or replenish RNF20 expression via intramuscular AAV injection. We found that blocking ID-induced ferritinophagy or up-regulating RNF20 expression could restore the differentiation and regeneration of skeletal muscle. To further understand the biological function of NCOA4 in SCs under ID conditions, we constructed NCOA4 cKO mice in SCs and found that reducing the expression of NCOA4 in SCs could inhibit the degradation of RNF20 and increase the level of H2Bub1 under ID conditions. Correspondingly, the skeletal muscle regeneration defect under ID conditions was restored in NCOA4SCs/KO mice compared to the control group. Moreover, a low iron level was accompanied by skeletal muscle regeneration impairment in iron-deficient individuals. Moreover, RNF20 and H2Bub1 expression levels in the skeletal muscle were down-regulated in the iron-deficient patient group.
As previously reported, iron supplementation rescued muscle mass and function in patients with cancer cachexia (12). Whether iron supplementation exhibited this effect through RNF20-H2Bbu1 axis is still need further investigation. Because of the negative effects of either iron overload or ID on skeletal muscle, precise regulation of muscle differentiation and regeneration in molecular mechanism level may be a better way for iron-deficient patients or elderly individuals. Here, our study provides insights into therapeutic targets that can be leveraged to increase muscle mass effectively in these patients via skeletal muscle regeneration mediated through selectively regulated ferritinophagy or directly by enhancing RNF20 expression and activity.
We found that ID inhibited muscle differentiation and regeneration by activating ferritinophagy. This process was conducted by the ferritinophagy-specific cargo protein NCOA4 binding to and degrading RNF20, which decreased the level of H2B monoubiquitination of the MyoG and MyoD myogenic markers. Our research had certain limitations, one of which was the in vitro mechanism study primarily using the C2C12 cell line rather than primary SCs. Disputation still remains, considering that C2C12 cell lines are markedly different from primary SCs and that using C2C12 cell lines to dissecting biology of myogenic differentiation and regeneration may result in discrepancies. However, we believe that the use of C2C12 cells instead of primary SCs for mechanistic investigations does not compromise the accuracy of our conclusions for the following reasons. From one hand, we initially validated the most crucial findings of this study in human primary SCs and C2C12 cells. These results were consistent to show that ID induced ferritinophagy, thereby impeding myogenic differentiation. From the other hand, the in vivo mouse experiments further supported our findings. Conditional knockout of NCOA4 in mouse SCs could reverse the decrease in RNF20 and H2Bub1 expression in mouse skeletal muscle under ID conditions, thereby restoring the regeneration capacity of mouse skeletal muscles. Therefore, we believe that ID induced ferritinophagy to mediate the degradation of RNF20 and thus suppress the H2Bub1 level is likely to be a universal phenomenon, rather than dependent on cell type such as C2C12. Further studies need to be performed to address this issue. Besides, we studied this phenomenon only in skeletal muscle cells, and it remains unclear whether this process contributes to other pathophysiology processes in other organs or systems. The present results provide insight into the pathological mechanism of different diseases caused by ID, which is the focus of our future experiments.
MATERIALS AND METHODS
Chemicals and reagents
The following commercially available reagents were used for this study: DFO (HY-B0988, MCE), DFP (HY-B0568, MCE), 3-MA (5142-23-4, Sigma-Aldrich), DC661 (D881024-2, Macklin), AFC (MB2808, Meilunbio), MG132 (T215-4, TargetMol), and CTX (217503, Merck Millipore). Information on the antibodies used in this study is shown in table S2.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of The Eighth Affiliated Hospital, Sun Yat-sen University. Written informed consent was obtained from all subjects included in the study in accordance with the Declaration of Helsinki. All animal experimental procedures were approved by the Animal Ethical and Welfare Committee of The Eighth Affiliated Hospital, Sun Yat-sen University.
Animals
C57BL/6 male mice were housed in the pathogen-free barrier facility of Sun Yat-sen University in accordance with institutional guidelines and approved by the regional board. Mice were maintained in a temperature- and humidity-controlled animal facility with a 12-hour light/12-hour dark cycle and free access to distilled water. The animals were kept in stainless steel mesh wire bottom cages with no bedding material and no access to feces.
C57BL/6 Pax7-CreER and C57BL/6 NCOA4fl/fl mice were purchased from the Jackson Laboratory. Mice with NCOA4-specific deletion in SCs were generated by crossing C57BL/6 Pax7-CreER and C57BL/6 NCOA4fl/fl mice. SC-specific NCOA4 deletion was induced by intraperitoneal injection of TMX (T5648, Sigma-Aldrich, St. Louis, MO) dissolved in corn oil for seven consecutive days at a dose of 15 mg/ml.
All mice were approximately 21 days of age on day 0 of treatment. The mice were given a control diet [37 parts per million (ppm) of Fe; Dyets, 110700] and an iron-deficient diet (approximately 1 ppm of Fe; Dyets, D115109) for 8 weeks, and they were weighed two to three times per week from the first day on the diets up to and including the day of sacrifice. For 3-MA and AAV overexpression assay in vivo, mice in the ID + 3-MA group received 3-MA (15 mg/kg per day) 2 weeks before euthanizing, and mice in the ID + AAV-RNF20 group received AAV-RNF20 4 weeks before euthanizing. CTX was used to induce skeletal muscle injury. Mice were anesthetized, and 50 μl of CTX [10 μM in phosphate-buffered saline (PBS)] was injected into the hindlimb GA muscles to induce injury. 3-MA or AAV-RNF20 was added as described above, respectively, 2 and 4 weeks before CTX injection. After 7 days of CTX injection, the mice were euthanized. GA muscle sections were subjected to further analyses.
Grip strength test
Grip strength was measured with an electronic dynamometer (HandpiHP-5N, China). Each mouse was positioned along a straight line parallel to the grip. As the pulling force gradually increased, when the mouse could not bear the force, it released the dynamometer, and the maximum pulling force was recorded. The forelimb grip strength of each mouse was tested three times, and the highest value was applied in the analysis.
Hanging grid test
In this assay, inverted hanging time was measured. A 45 cm–by–45 cm grid was placed on a 55-cm-high frame, and a 5-cm-thick cushion was placed under the grid. We placed each mouse at the center of the grid and then turned the grid upside down. Hanging time was recorded as the time after which the mouse fell. Each mouse was tested three times with a >30-min interval between tests, and the highest value was applied in the analysis.
Exhaustive running test
Exhaustive running time was measured using a treadmill. The mouse began running at 13 m/min with an inclination of 0°, and the speed and inclination were then increased by 2 m/min and 2°, respectively, every 3 min until they reached 39 m/min and 14°, respectively. The mice were considered exhausted and removed from the treadmill when the mouse being tested did not return to the track for >20 s and concomitantly exhibited a markedly diminished response to external stimuli. The exhaustive running time was recorded.
Culture of C2C12 cells
C2C12 cells were obtained from the Chinese Academy of Sciences Cell Bank and grown in incubators at 37°C and 5% CO2, and proliferating cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco, Australia). When C2C12 cells reached 80 to 90% confluence, they were digested with 0.25% trypsin containing 0.53 mM EDTA and reseeded in new flasks. To induce differentiation, the C2C12 cells were cultured in high-glucose DMEM supplemented with 2% horse serum.
Isolation and culture of SCs
Human MuSCs were isolated from human biopsies as CD31−/CD45−/CD56+ cells, and MuSCs were isolated from mice as CD31−/CD45−/Scal−/α7-integrin+ cells. Briefly, human biopsies or hindlimb muscles for each mouse were carefully dissected to remove attached tendons, nerves, blood vessels, and fat tissue. Muscles were dissociated mechanically and digested in bovine serum albumin (BSA; Sigma-Aldrich), penicillin (10 U/ml) and streptomycin (10 μg/ml), collagenase A (2 mg/ml) (Roche), and dispase II (2.4 U/ml; Roche) in Hanks’ balanced buffer solution (Gibco) for 90 min at 37°C. The supernatants were filtered through 70- and 40-μm cell strainers. Erythrocytes were eliminated by treating the cells with red blood cell lysis solution.
The Miltenyi MACS purification system was used to isolate MuSCs. Briefly, the cells isolated from digested muscles were resuspended in MACS buffer consisting of PBS with 2% FBS and 2 mM EDTA and incubated with FcR blocking reagent (Miltenyi Biotec) for 10 min at 4°C, followed by incubation with biotin-conjugated anti-CD31, anti-CD45, anti-CD56, anti-sca1 and anti–α7-integrin (BioLegend) for 20 min at 4°C. Subsequently, the cells were washed once with MACS buffer and incubated with anti-biotin microbeads (Miltenyi Biotec). Antibody-microbead cellular complexes were passed through a magnetic LD column, and the flow through the fraction was collected. The following populations were MACS isolated: CD31−/CD45−/CD56+ cells (MuSCs in human biopsies) and CD31−/CD45−/Scal−/α7-integrin+ cells (MuSCs in mice). Cells were cultured in growth medium (DMEM supplemented with 20% FBS) at 37°C and 5% CO2. To induce differentiation, the cells were cultured in DMEM supplemented with 5% horse serum.
Single myofiber isolation
Extensor digitorum longus muscles were isolated and incubated with 0.2% type I collagenase (Sigma-Aldrich) in DMEM for 1 hour. After digestion, single myofibers were transferred to 24-well plates and fixed with 4% paraformaldehyde for subsequent immunostaining.
Cell counting kit-8 assay
A total of 1 × 104 C2C12 cells per well were seeded for use in a cytotoxicity assay, and 2 × 103 C2C12 cells per well were seeded for use in a proliferation assay. At each measurement time point, fresh culture medium with Cell counting kit-8 reagent was added (Dojindo, Japan). After 2 hours of cell culture, the absorbance of the wells was measured at 450 nm. Medium without cells was used as negative controls.
Western blot analysis and antibodies
Western blot protocols have been previously reported (54). Briefly, cells and tissues were lysed in radioimmunoprecipitation assay buffer containing protease and phosphatase inhibitors for 30 min on ice, and lysates were collected and centrifuged at 14,000 rpm at 4°C for 10 min. Protein lysates were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred to polyvinylidene fluoride membranes (Millipore). After blocking in 5% nonfat dry milk dissolved in TBST [150 mM NaCl, 50 mM tris-HCl (pH of 7.5), and 0.05% Tween 20] at room temperature for 1 hour, the membranes were incubated with primary antibodies overnight at 4°C. After three washes in TBST, the membranes were incubated with horseradish peroxidase–conjugated AffiniPure secondary antibodies (1:5000; Boster, BA1054 and BA1050) for 1 hour at room temperature and were detected using chemiluminescent reagents (Millipore) according to the manufacturer’s instructions.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed as previously described (55). Briefly, total RNA was isolated from cells using an RNA Quick Purification kit (ESscience) according to the manufacturer’s instructions and was transcribed into cDNA using a PrimeScript RT reagent kit (TaKaRa). qRT-PCR was then performed on a Light-Cycler 480 PCR System (Roche) using SYBR Premix Ex Taq (TaKaRa). Gene expression was calculated by the ΔΔCT method using glyceraldehyde-3-phosphate dehydrogenase as the reference gene. The primer sequences used for qRT-PCR are shown in table S3.
Plasmid and lentivirus construction and transfection
Expression plasmid constructs, including full-length pcDNA3.1(+)-Flag-RNF20 [Mus musculus (house mouse)] and full-length pcDNA3.1(+)-Myc-NCOA4 (M. musculus) were all constructed and purchased from Guangzhou IGE Biotechnology, Ltd. Lipofectamine 3000 transfection reagent (Invitrogen, L3000-015) was used for transfection according to the manufacturer’s instructions with minor modifications. Briefly, 293T cells were seeded in six-well plates at a density of 2 × 105 cells per well. Cells were transfected with plasmids (2.5 μg per well), Lipofectamine 3000 (5 μl) and P3000 (5 μl) according to the manufacturer’s instructions. The PLenti-EF1a-EGFP-P2A-Puro-CMV- RNF20-3Flag construct was generated by Obio Technology Corp. Ltd. (Shanghai). C2C12 cells were incubated with lentiviruses for 12 hours at a multiplicity of infection of 50. The siRNA sequences are shown in table S4.
Recombinant AAV
pAAV-CMV-3xFLAG-Rnf20-tWPA [AAV2/9, 1.0 × 1013 viral genomes (V.G.)/ml] was generated by the Obio Technology Corp. Ltd. (Shanghai) and was used to overexpress RNF20 in mice. A single dose of AAV-RNF20 (2.5 × 1011 V.G./25 μl per point) was delivered to iron-deficient mice via GA muscle injection. The contralateral GA muscle was used as the control. Mice were evaluated for 4 weeks after treatment and then euthanized, and the GA muscle was harvested.
Co-IP and LC-MS/MS
C2C12 or 293T cells were quickly harvested and lysed on ice in Western and IP lysis buffer (P0013, Beyotime Biotechnology) containing 1% phenylmethylsulfonyl fluoride. Cell extracts (approximately 200 μg of total protein) were incubated with antibodies at 4°C overnight. Protein G Dynabeads (10007D, Thermo Fisher Scientific) were then added, and the mixture was incubated at 4°C for 3 hours. Dynabeads were collected, washed, and resuspended in 60 μl of sample buffer, and samples were then boiled for 10 min. SDS-PAGE was used to separate the samples, and a Coomassie blue staining kit (ESscience) was subsequently used. Differential beads were collected for further LC-MS/MS to analyze the interacting proteins of NCOA4 in C2C12 cells. Western blot analysis was performed using the abovementioned protocols. A unique secondary antibody (1:1000; Abcam, ab131366), which only recognizes native (nonreduced) antibodies to minimize the detection of heavy and light chains, was used to test the IP samples to prevent the influence of the IP antibodies in the IP samples.
ChIP-qPCR assays
The ChIP-qPCR assay was performed using a ChIP assay kit (Millipore, 17-10086) according to the manufacturer’s protocol with modifications. Overall, 5 mg of antibodies was used for each 1 × 107 cells in one IP experiment. Data are expressed as the percentage of input DNA. The following antibodies were used in the ChIP assays: anti-RNF20 (Cell Signaling Technology, 11974) and anti-H2Bub1 (Cell Signaling Technology, 5546S). Immunoprecipitated genomic DNA was collected with 50 μl of elution buffer. qRT-PCR was performed as described above. The primer sequences used for ChIP-PCR are shown in table S3.
LA-ICP-MS and in situ imaging analysis
For sample preparation, skeletal muscle samples were dried in a vacuum for 24 hours followed by epoxy resin coating. After microtome slicing, the prepared sample was subjected to laser ablation on a NRW 193HE instrument. For tissue imaging, the spot size and frequency of the laser were set to 15 μm by 15 μm and 10 Hz with a movement rate of 40 μm/s. The amount of trace elemental iron was calibrated on the basis of that in external NIST 610 glass level. An Agilent 8900 ICP-MS Triple Qua instrument was used to acquire ion signal intensities. Iolite software was used to perform off-line selection, background integration, signal analysis, time drift correction, and quantitative calibration for the trace element analysis.
H&E staining
Freshly isolated skeletal muscle was fixed with 4% polyoxymethylene for 24 hours and then embedded in paraffin for sectioning. After the slides were dewaxed and hydrated using dimethylbenzene and a gradient ethanol series, they were stained with hematoxylin for 5 min. After washing with PBS for 10 min, the sections were stained with eosin for 3 min. After dehydration with ethanol and clearing in a dimethylbenzene solution, the slides were mounted and observed using a microscope.
Prussian blue iron staining
The tissue was cut into paraffin sections of 3 to 7 μm. The slices were dewaxed to water routinely. Then, we use the Prussian Blue Iron Staining Kit (enhance with 3,3’-diaminobenzidine) (Solarbio, G1428) to stain non-heme iron in the skeletal muscle according to the kit’s protocol.
Immunofluorescence
The tissue was cut into paraffin sections of 3 to 7 μm. The slices were dewaxed to water routinely. Antigens were retrieved using an EDTA buffer in a microwave oven for 15 min. To permeabilize the sections, 0.1% Triton X-100 was added for 15 min at room temperature. Goat serum (10%) was added to block nonspecific binding. After the sections were washed with PBS, they were incubated with primary antibodies at 4°C overnight. The concentrations and sources of the antibodies that were used are listed below. After the sections were washed three times with PBS, anti-rabbit immunoglobulin G (IgG) (1:500; Cell Signaling Technology, 4412) and anti-mouse IgG (1:500; Cell Signaling Technology, 4409) were added, and the sections were incubated for another 1 hour at room temperature. Then, 4′,6-diamidino-2-phenylindole was used to counterstain the nuclei. Thereafter, the coverslips were mounted on glass slides with antifade mounting medium (Beyotime, P0131).
C2C12 cells were seeded on sterile glass coverslips. After induction of myogenic differentiation for 3 days, the growth medium was removed. After three washes with PBS for 5 min each time, the cells were fixed, permeabilized, blocked, and processed for incubation with the primary antibodies described above overnight at 4°C, followed by secondary staining and counterstaining of the nuclei. Thereafter, the samples were observed under a laser scanning confocal microscope (LSM 880 with Airyscan) at wavelengths of 488 nm (green), 561 nm (red), and 405 nm (blue).
Human skeletal muscle study
The participants assessed in this study were recruited from The Eighth Affiliated Hospital, Sun Yat-sen University. Each of the control and iron-deficient groups consisted of six age- and sex-matched subjects. The study included patients admitted to the hospital for orthopedic surgery on fractures. Serum samples from the participants (patients with ID and healthy controls) were obtained and measured in the clinic laboratory. Hemoglobin and hematocrit levels were provided by the Department of Laboratory Medicine, The Eighth Affiliated Hospital of Sun Yat-sen University. Serum iron levels and total iron-binding capacity were quantified using a total iron binding capacity (TIBC) and the Serum Iron Assay Kit (ab239715). Transferrin saturation was calculated as follows: transferrin saturation (%) = serum iron/(TIBC) × 100.
Statistical analysis
All experiments included at least three biological replicates. The areas of fluorescent staining for muscle fibers and nuclei were determined using ImageJ software. All data are expressed as the means ± SD. The unpaired Student’s t test was used for comparisons between two groups. For comparison of more than two groups, one-way analysis of variance (ANOVA) was used. Data were analyzed using GraphPad Prism 8.0 software. *P < 0.05 and **P < 0.01 indicate statistical significance.
Acknowledgments
We would like to thank the animal facility at Sun Yat-sen University. C2C12 cells were kindly provided by the Chinese Academy of Sciences Cell Bank. The entire LC-MS/MS procedure was performed by the Bioinformatics and Omics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
Funding: This study was financially supported by the National Natural Science Foundation of China (81971518 and 82002267), the Key-Area Research and Development Program of Guangdong Province (2019B020236001), the Shenzhen Key Laboratory of Ankylosing Spondylitis (ZDSYS20190902092851024), the Basic and Applied Basic Research Fund of Guangdong Province (2020A1515010097), and the Science and Technology Project of Shenzhen City (JCYJ20190808101005694).
Author contributions Z.X., J. Li, P.W., and Y.C. conceived the study, generated hypotheses, and designed experiments. P.X. and Z.Z. collected the human samples. Y.C., W.Y., J. Lin, Z.S., and F.Y. performed the experiments and analyzed the data. Y.C., J Li, and Z.X. wrote, reviewed, and edited the paper. Y.W. and H.S. supervised the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S5
REFERENCES AND NOTES
- 1.R. J. Ward, R. R. Crichton, D. L. Taylor, L. D. Corte, S. K. Srai, D. T. Dexter, Iron and the immune system. J. Neural Transm. 118, 315–328 (2011). [DOI] [PubMed] [Google Scholar]
- 2.R. A. Weber, F. S. Yen, S. P. V. Nicholson, H. Alwaseem, E. C. Bayraktar, M. Alam, R. C. Timson, K. La, M. Abu-Remaileh, H. Molina, K. Birsoy, Maintaining iron homeostasis is the key role of lysosomal acidity for cell proliferation. Mol. Cell 77, 645–655.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. J. Anderson, C. D. Vulpe, Mammalian iron transport. Cell. Mol. Life Sci. 66, 3241–3261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. J. Ward, F. A. Zucca, J. H. Duyn, R. R. Crichton, L. Zecca, The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13, 1045–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.K. Roemhild, F. von Maltzahn, R. Weiskirchen, R. Knüchel, S. von Stillfried, T. Lammers, Iron metabolism: Pathophysiology and pharmacology. Trends Pharmacol. Sci. 42, 640–656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Y. Jiang, C. Li, Q. Wu, P. An, L. Huang, J. Wang, C. Chen, X. Chen, F. Zhang, L. Ma, S. Liu, H. He, S. Xie, Y. Sun, H. Liu, Y. Zhan, Y. Tao, Z. Liu, X. Sun, Y. Hu, Q. Wang, D. Ye, J. Zhang, S. Zou, Y. Wang, G. Wei, Y. Liu, Y. Shi, Y. Eugene Chin, Y. Hao, F. Wang, X. Zhang, Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat. Commun. 10, 2935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.G. F. Allen, R. Toth, J. James, I. G. Ganley, Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 14, 1127–1135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. Pasricha, J. Tye-Din, M. U. Muckenthaler, D. W. Swinkels, Iron deficiency. Lancet 397, 233–248 (2021). [DOI] [PubMed] [Google Scholar]
- 9.X. Gao, Y. Song, J. Wu, S. Lu, X. Min, L. Liu, L. Hu, M. Zheng, P. Du, Y. Yu, H. Long, H. Wu, S. Jia, D. Yu, Q. Lu, M. Zhao, Iron-dependent epigenetic modulation promotes pathogenic T cell differentiation in lupus. J. Clin. Invest. 132, e152345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.G. Corna, I. Caserta, A. Monno, P. Apostoli, A. A. Manfredi, C. Camaschella, P. Rovere-Querini, The repair of skeletal muscle requires iron recycling through macrophage ferroportin. J. Immunol. 197, 1914–1925 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Y. Ikeda, A. Satoh, Y. Horinouchi, H. Hamano, H. Watanabe, M. Imao, M. Imanishi, Y. Zamami, K. Takechi, Y. Izawa Ishizawa, L. Miyamoto, T. Hirayama, H. Nagasawa, K. Ishizawa, K. I. Aihara, K. Tsuchiya, T. Tamaki, Iron accumulation causes impaired myogenesis correlated with MAPK signaling pathway inhibition by oxidative stress. FASEB J. 33, 9551–9564 (2019). [DOI] [PubMed] [Google Scholar]
- 12.E. Wyart, M. Y. Hsu, R. Sartori, E. Mina, V. Rausch, E. S. Pierobon, M. Mezzanotte, C. Pezzini, L. B. Bindels, A. Lauria, F. Penna, E. Hirsch, M. Martini, M. Mazzone, A. Roetto, S. Geninatti Crich, H. Prenen, M. Sandri, A. Menga, P. E. Porporato, Iron supplementation is sufficient to rescue skeletal muscle mass and function in cancer cachexia. EMBO Rep. 23, e53746 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.F. M. Alves, K. Kysenius, M. K. Caldow, J. P. Hardee, P. J. Crouch, S. Ayton, A. I. Bush, G. S. Lynch, R. Koopman, Iron accumulation in skeletal muscles of old mice is associated with impaired regeneration after ischaemia–reperfusion damage. J. Cachexia. Sarcopenia Muscle 12, 476–492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. Dziegala, K. Josiak, M. Kasztura, K. Kobak, S. von Haehling, W. Banasiak, S. D. Anker, P. Ponikowski, E. Jankowska, Iron deficiency as energetic insult to skeletal muscle in chronic diseases. J. Cachexia. Sarcopenia Muscle 9, 802–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.J. F. Merrill, D. M. Thomson, S. E. Hardman, S. D. Hepworth, S. Willie, C. R. Hancock, Iron deficiency causes a shift in AMP-activated protein kinase (AMPK) subunit composition in rat skeletal muscle. Nutr. Metab. 9, 104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N. Mizushima, A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527 (2018). [DOI] [PubMed] [Google Scholar]
- 17.D. C. Rubinsztein, P. Codogno, B. Levine, Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. F. Bento, M. Renna, G. Ghislat, C. Puri, A. Ashkenazi, M. Vicinanza, F. M. Menzies, D. C. Rubinsztein, Mammalian autophagy: How does it work? Annu. Rev. Biochem. 85, 685–713 (2016). [DOI] [PubMed] [Google Scholar]
- 19.J. H. Hurley, L. N. Young, Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86, 225–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Kaushik, A. M. Cuervo, Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. Lee, F. Wilfling, P. Ronchi, M. Allegretti, S. Mosalaganti, S. Jentsch, M. Beck, B. Pfander, Selective autophagy degrades nuclear pore complexes. Nat. Cell Biol. 22, 159–166 (2020). [DOI] [PubMed] [Google Scholar]
- 22.K. A. Bauckman, N. Owusu-Boaitey, I. U. Mysorekar, Selective autophagy: Xenophagy. Methods 75, 120–127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. Bellelli, G. Federico, A. Matte, D. Colecchia, A. Iolascon, M. Chiariello, M. Santoro, L. De Franceschi, F. Carlomagno, NCOA4 deficiency impairs systemic iron homeostasis. Cell Rep. 14, 411–421 (2016). [DOI] [PubMed] [Google Scholar]
- 24.A. Nai, M. R. Lidonnici, G. Federico, M. Pettinato, V. Olivari, F. Carrillo, C. S. Geninatti, G. Ferrari, C. Camaschella, L. Silvestri, F. Carlomagno, NCOA4-mediated ferritinophagy in macrophages is crucial to sustain erythropoiesis in mice. Haematologica 106, 795–805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. Quiles Del Rey, J. D. Mancias, NCOA4-mediated ferritinophagy: A potential link to neurodegeneration. Front. Neurosci. 13, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. D. Mancias, X. Wang, S. P. Gygi, J. W. Harper, A. C. Kimmelman, Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.N. Santana-Codina, J. Mancias, The role of NCOA4-mediated ferritinophagy in health and disease. Pharmaceuticals 11, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. J. Marsh, Y. Ma, K. Dickson, Histone monoubiquitination in chromatin remodelling: Focus on the histone H2B interactome and cancer. Cancers 12, 3462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.X. Liang, C. Tao, J. Pan, L. Zhang, L. Liu, Y. Zhao, Y. Fan, C. Cao, J. Liu, J. Zhang, S. M. Lam, G. Shui, W. Jin, W. Li, J. Zhao, K. Li, Y. Wang, Rnf20 deficiency in adipocyte impairs adipose tissue development and thermogenesis. Protein Cell 12, 475–492 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. Chen, Y. Jing, X. Kang, L. Yang, D. Wang, W. Zhang, L. Zhang, P. Chen, J. Chang, X. Yang, F. Sun, Histone H2B monoubiquitination is a critical epigenetic switch for the regulation of autophagy. Nucleic Acids Res. 45, 1144–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B. Lee, J. Lee, W. Shen, C. Rhee, H. Chung, J. Kim, Fbxl19 recruitment to CpG islands is required for Rnf20-mediated H2B mono-ubiquitination. Nucleic Acids Res. 45, 7151–7166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.G. Fuchs, E. Shema, R. Vesterman, E. Kotler, Z. Wolchinsky, S. Wilder, L. Golomb, A. Pribluda, F. Zhang, M. Haj-Yahya, E. Feldmesser, A. Brik, X. Yu, J. Hanna, D. Aberdam, E. Domany, M. Oren, RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol. Cell 46, 662–673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.G. Fuchs, M. Oren, Writing and reading H2B monoubiquitylation. Biochim. Biophys. Acta 1839, 694–701 (2014). [DOI] [PubMed] [Google Scholar]
- 34.F. Lai, H. Wang, X. Zhao, K. Yang, L. Cai, M. Hu, L. Lin, X. Xia, W. Li, H. Cheng, R. Zhou, RNF20 is required for male fertility through regulation of H2B ubiquitination in the Sertoli cells. Cell Biosci. 13, 71 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.G. Sethi, M. K. Shanmugam, F. Arfuso, A. P. Kumar, Role of RNF20 in cancer development and progression - a comprehensive review. Biosci. Rep. 38, BSR20171287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O. Tarcic, I. S. Pateras, T. Cooks, E. Shema, J. Kanterman, H. Ashkenazi, H. Boocholez, A. Hubert, R. Rotkopf, M. Baniyash, E. Pikarsky, V. G. Gorgoulis, M. Oren, RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep. 14, 1462–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Qi, Y. Liu, J. Zhai, Y. Wang, W. Li, T. Wang, Y. Chai, RNF20 deletion causes inflammation in model of sepsis through the NLRP3 activation. Immunopharmacol. Immunotoxicol. 45, 469–478 (2023). [DOI] [PubMed] [Google Scholar]
- 38.S. Chen, J. Li, D. Wang, F. Sun, Histone H2B lysine 120 monoubiquitination is required for embryonic stem cell differentiation. Cell Res. 22, 1402–1405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. Wu, L. Li, B. Zhou, Z. Qin, Y. Dou, H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol. Cell 54, 920–931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.F. M. Alves, K. Kysenius, M. K. Caldow, J. P. Hardee, J. D. Chung, J. Trieu, D. J. Hare, P. J. Crouch, S. Ayton, A. I. Bush, G. S. Lynch, R. Koopman, Iron overload and impaired iron handling contribute to the dystrophic pathology in models of Duchenne muscular dystrophy. J. Cachexia. Sarcopenia Muscle 13, 1541–1553 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Stugiewicz, M. Tkaczyszyn, M. Kasztura, W. Banasiak, P. Ponikowski, E. A. Jankowska, The influence of iron deficiency on the functioning of skeletal muscles: Experimental evidence and clinical implications. Eur. J. Heart Fail. 18, 762–773 (2016). [DOI] [PubMed] [Google Scholar]
- 42.K. Kobak, M. Kasztura, M. Dziegala, J. Bania, V. Kapuśniak, W. Banasiak, P. Ponikowski, E. A. Jankowska, Iron limitation promotes the atrophy of skeletal myocytes, whereas iron supplementation prevents this process in the hypoxic conditions. Int. J. Mol. Med. 41, 2678–2686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Y. Tian, J. Lu, X. Hao, H. Li, G. Zhang, X. Liu, X. Li, C. Zhao, W. Kuang, D. Chen, M. Zhu, FTH1 inhibits ferroptosis through ferritinophagy in the 6-OHDA model of parkinson’s disease. Neurotherapeutics 17, 1796–1812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Q. Yan, W. Zhang, M. Lin, O. Teymournejad, K. Budachetri, J. Lakritz, Y. Rikihisa, Iron robbery by intracellular pathogen via bacterial effector–induced ferritinophagy. Proc. Natl. Acad. Sci. U. S. A. 118, e2026598118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K. A. Bauckman, I. U. Mysorekar, Ferritinophagy drives uropathogenicEscherichia coli persistence in bladder epithelial cells. Autophagy 12, 850–863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Goodall, A. Thorburn, Identifying specific receptors for cargo-mediated autophagy. Cell Res. 24, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. A. Call, R. J. Wilson, R. C. Laker, M. Zhang, M. Kundu, Z. Yan, Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am. J. Physiol. Cell Physiol. 312, C724–C732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. Sin, A. M. Andres, D. J. R. Taylor, T. Weston, Y. Hiraumi, A. Stotland, B. J. Kim, C. Huang, K. S. Doran, R. A. Gottlieb, Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 12, 369–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.C. H. Kim, K. H. Kim, Y. Yoo, Melatonin-induced autophagy is associated with degradation of MyoD protein in C2C12 myoblast cells. J. Pineal Res. 53, 289–297 (2012). [DOI] [PubMed] [Google Scholar]
- 50.E. M. McMillan, J. Quadrilatero, Autophagy is required and protects against apoptosis during myoblast differentiation. Biochem. J. 462, 267–277 (2014). [DOI] [PubMed] [Google Scholar]
- 51.S. Thapaliya, A. Runkana, M. R. McMullen, L. E. Nagy, C. McDonald, S. V. N. Prasad, S. Dasarathy, Alcohol-induced autophagy contributes to loss in skeletal muscle mass. Autophagy 10, 677–690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Q. Liang, W. Xia, W. Li, J. Jiao, RNF20 controls astrocytic differentiation through epigenetic regulation of STAT3 in the developing brain. Cell Death Differ. 25, 294–306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Y. Ishino, Y. Hayashi, M. Naruse, K. Tomita, M. Sanbo, T. Fuchigami, R. Fujiki, K. Hirose, Y. Toyooka, T. Fujimori, K. Ikenaka, S. Hitoshi, Bre1a, a histone H2B ubiquitin ligase, regulates the cell cycle and differentiation of neural precursor cells. J. Neurosci. 34, 3067–3078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. Li, P. Wang, Z. Xie, S. Wang, S. Cen, M. Li, W. Liu, S. A. Tang, G. Ye, G. Zheng, H. Su, M. Ma, X. Wu, Y. Wu, H. Shen, TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 26, 2652–2666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.G. Ye, Z. Xie, H. Zeng, P. Wang, J. Li, G. Zheng, S. Wang, Q. Cao, M. Li, W. Liu, S. Cen, Z. Li, Y. Wu, Z. Ye, H. Shen, Oxidative stress-mediated mitochondrial dysfunction facilitates mesenchymal stem cell senescence in ankylosing spondylitis. Cell Death Dis. 11, 775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S5