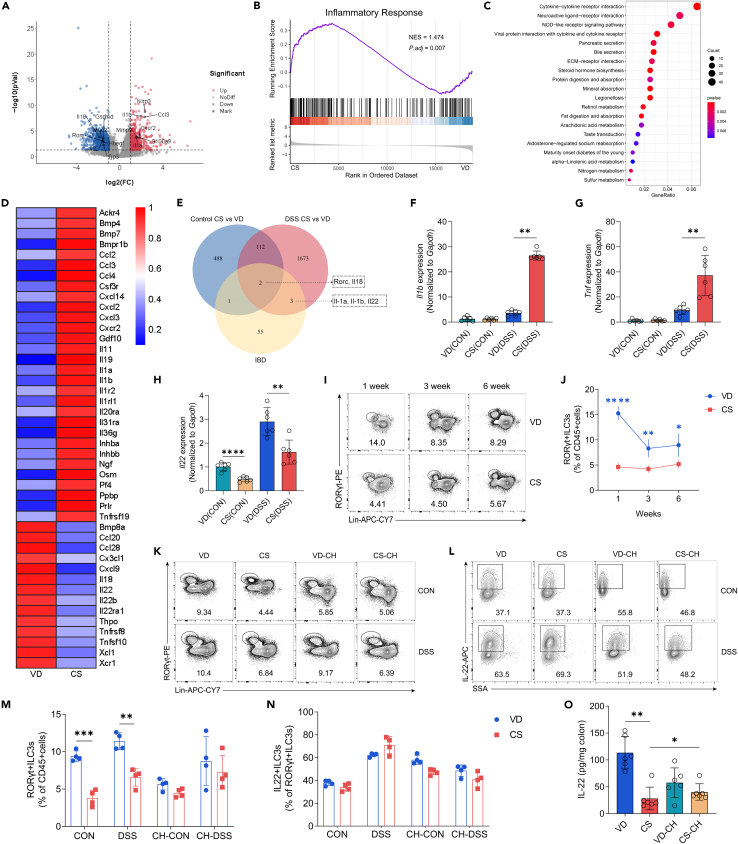
Figure 3.
CS impairs ILC3 development and IL-22 expression in the intestine
(A) Volcano plot showing the significantly different up-regulated and down-regulated genes of colon tissues in CS (DSS) group compared with those in VD (DSS) group (n = 4 per group).
(B) GSEA of RNA-seq data revealed gene sets significantly enriched in CS (DSS) mice compared with VD (DSS) mice, particularly for gene sets involved in the inflammatory response signaling pathway (n = 4 per group).
(C) Bubble plot showing the top 20 enriched KEGG pathways for DEGs between VD (DSS) and CS (DSS) group (n = 4 per group).
(D) Heatmap showing expression levels of DEGs between VD (DSS) and CS (DSS) group in the list of “Cytokine-cytokine receptor pathway” term in the KEGG database (n = 4 per group).
(E) Venn diagram illustrating the number of shared DEGs among the 3 gene sets (n = 4 per group). Control CS vs. VD: DEGs between CS (CON) and VD (CON); DSS CS vs. VD: DEGs between CS (DSS) and VD (DSS); IBD: Genes in the list of “inflammatory bowel disease” term in the KEGG database.
(F–H) qRT-PCR analysis of relative Il1b (F), Tnf (G) and Il22 (H) expression in the colon tissue of VD and CS mice from untreated control group or 3% DSS-treated group (n = 6 per group).
(I and J) Representative flow cytometry plots and percentage of colonic ILC3s (liveCD45+Lin-RORγt+) from VD and CS mice at defined ages (n = 4 per group).
(K–N) Representative flow cytometry plots and percentage of colonic ILC3s (K and M) and IL-22+ILC3s (L and N) from adult VD, CS, co-housed VD and co-housed CS mice at steady state or after exposure to 3% DSS (n = 4 per group).
(O) ELISA determined the protein levels of IL-22 in the colon tissue of VD, CS, co-housed VD and co-housed CS mice treated with 3% DSS (n = 6 per group).
Data shown as the mean ± SEM. One dot represents one mouse. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.