Abstract
Purpose
Providing feasible preimplantation genetic testing strategies for monogenic disorders (PGT-M) for prevention and control of genetic cancers.
Methods
Inclusion of families with a specific pathogenic mutation or a clear family history of genetic cancers. Identification of the distribution of hereditary cancer-related mutations in families through genetic testing. After a series of assisted reproductive measures such as down-regulation, stimulation, egg retrieval, and in vitro fertilization, a biopsy of trophectoderm cells from a blastocyst was performed for single-cell level whole-genome amplification (WGA). Then, the detection of chromosomal aneuploidies was performed by karyomapping. Construction of a haplotype-based linkage analysis to determine whether the embryo carries the mutation. Meanwhile, we performed CNV testing. Finally, embryos can be selected for transfer, and the results will be verified in 18–22 weeks after pregnancy.
Results
Six couples with a total of 7 cycles were included in our study. Except for cycle 1 of case 5 which did not result in a transferable embryo, the remaining 6 cycles produced transferable embryos and had a successful pregnancy. Four couples have had amniotic fluid tests to confirm that the fetus does not carry the mutation, while 1 couple was not tested due to insufficient pregnancy weeks. And the remaining couples had to induce labor due to fetal megacystis during pregnancy.
Conclusion
Our strategy has been proven to be feasible. It can effectively prevent transmission of hereditary cancer-related mutations to offspring during the prenatal stage.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-023-02939-0.
Keywords: Preimplantation genetic testing for monogenic disorders, Karyomapping, Hereditary cancers, Haplotype linkage analysis
Introduction
Tumors are currently one of the major health threats in the world. Over the past few decades, the incidence of tumors has been on the rise, both in developed and developing countries [1, 2]. Besides, the mortality rate associated with cancer has also been increasing. While treatments have improved, the mortality rate remains high [2]. Hereditary tumor is one of the major types of malignant tumors. Approximately 5–10% of all cancers are caused by inherited mutations in tumor susceptibility genes [3–5]. Inherited cancer-related mutations can significantly increase the risk for associated cancer and pose major challenges to the prevention and treatment of tumors. Hereditary tumors are common in a variety of tumors. Take breast cancer as an example, breast cancer is currently the first tumor (31%) in the number of new cases and the second (15%) in mortality in women [1], and the new study is the first to document an elevated risk of breast cancer recurrence (BCR) persisting more than 25 years after primary surgery [6]. However, 5–10% of breast cancer patients are associated with inherited cancer-associated genes [7]. These figures indicate the urgent need for effective interventions to prevent and control tumors, especially hereditary tumors.
Preimplantation genetic testing (PGT) is a rapidly evolving technology that is used to diagnose genetic disorders in embryos before they are implanted into the uterus [8]. The technology has advanced significantly in recent years and is now widely used for a variety of purposes, including detecting single-gene disorders, chromosomal abnormalities, and other genetic conditions [9, 10]. PGT-M currently adopts the strategy of constructing haploid, with linkage analysis of single nucleotide polymorphisms (SNPs) located upstream and downstream of the pathogenic or predisposing genes [11–13]. The strategy of linkage analysis can reduce the risks and serious consequences of allele dropout (ADO) [14, 15].
The current prevention and treatment strategy for hereditary tumors is to conduct regular prevention and physical examinations for family members with a family history of hereditary tumors or to perform prenatal diagnosis during pregnancy [16]. Cancer in a member will increase the financial and psychological burden of the family [17], and the induction of labor if abnormalities are found in the prenatal stage will cause great harm to the pregnant woman's body [18].
PGT-M is a medically and ethically acceptable way to reduce the considerable risk of every offspring to inherit a familial tumor-predisposing mutation, and some studies have been conducted on PGT-M in hereditary tumor families [19–22]. The Ethics Taskforce of the European Society of Human Reproduction and Embryology declared that it is acceptable to perform PGT-M for late-onset and multifactorial diseases in 2003 [23]. In this study, we summarized and combed through all pedigrees with hereditary tumor family history in our center and aimed to explore the application of our technology in blocking the transmission of hereditary cancer-related mutations to offspring during the preconception period. This study involved hereditary tumors such as breast cancer, familial medullary thyroid carcinoma, renal cell carcinoma, retinoblastoma, and various hereditary tumors caused by the tumor susceptibility gene p53. All six families have successfully blocked familial cancer-predisposing genes and obtained pregnant or live births, demonstrating the feasibility of our strategy.
Materials and methods
Patients
Six couples suffering from hereditary tumors from the beginning of the Center of Reproductive Medicine of the First Affiliated Hospital of Zhengzhou University to April 1, 2023, were enrolled in this study. All the families of hereditary tumors included had a specific pathogenic mutation or a clear family history. Characteristics of including couples are shown in Table 1. All couples in the pedigree included have signed an informed consent form at genetic counseling. The subject was approved by the Internal Review Board of The First Affiliated Hospital of Zhengzhou University (Ethic no. 2023-KY-0361), and written informed consent was obtained from all patients.
Table 1.
Characteristics of the included couples
| Case | Male age | Male karyotype | Female age | Female karyotype | Hereditary cancer | Hereditary cancer-associated genes/affected partner/ACMG† | Reference | Reference status | Cycle | Retrieved oocytes | MII oocytes | 2PN | Biopsied blastocysts |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | 46,XY | 31 | 46,XX | Medullary thyroid carcinoma | RET/female/P | Daughter | RET(+) | 1 | 23 | 23 | 14 | 9 |
| 2 | 28 | 46,XY,21pstk+ | 30 | 46,XX | Breast cancer | BRCA1/male/P | Father | BRCA1(+) | 1 | 18 | 15 | 11 | 7 |
| 3 | 36 | 46,XY | 30 | 46,XX | Multiple tumors (tumor susceptibility genes) |
RAD51D/male/LP TP53/male/LP |
Son |
RAD51D(+) TP53(+) |
1 | 14 | 9 | 7 | 7 |
| 4 | 29 | 46,XY | 33 | 46,XX | Retinoblastoma | RB1/male/LP | Amniotic fluid | RB1(+) | 1 | 15 | 11 | 10 | 3 |
| 5 | 44 | 46,XY | 44 | 46,XX | Renal cell carcinoma | FH/male/VOUS | Daughter | FH(+) | 1 | 18 | 17 | 13 | 4 |
| 44 | 46,XY | 44 | 46,XX | Renal cell carcinoma | FH/male/VOUS | Daughter | FH(+) | 2 | 23 | 19 | 12 | 8 | |
| 6 | 36 | 46,XY | 37 | 46,XX | Breast cancer | BRCA1/male/P | Daughter | BRCA1(-) | 1 | 15 | 9 | 6 | 5 |
†P, pathogenic; LP, likely pathogenic; VOUS, variants of uncertain significance
Workflow of analysis
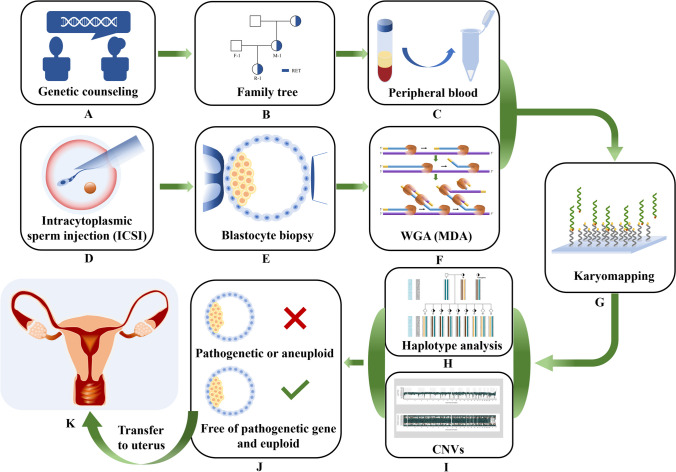
The flow of the method is shown in detail in Fig. 1. First, genetic counseling and peripheral blood collection were used to determine the hereditary cancer-related mutation and their distribution in the family. At the same time, the fertilization process is completed by intracytoplasmic sperm injection (ICSI), and the biopsy of the trophectoderm (TE) cell of the resulting embryo was performed to whole-genome amplification (WGA). Next, the copy number variations (CNVs) and the SNP flanking of the mutation were obtained by karyomapping technology. CNV was applied to detect aneuploidy of the embryo, and SNP information was used for linkage analysis. After SNP linkage analysis, haplotypes can be constructed based on references of the pedigree to assess the carrier status of all embryos. Then, non-carrier embryos can be selected to be transferred into the uterus. And we can complete the transmission of risk genes associated with hereditary tumors to offspring.
Fig. 1.
Workflow of analysis. (A–C) The main purpose is to obtain family information through genetic counseling and to clarify the mutated genes and their distribution in the family. (D–F) This is an embryo biopsy and preimplantation genetic testing. (G) This means that we obtain SNP information and CNV results through the karymapping chip. (H) It shows that we selected SNPs with 2M SNP upstream and downstream of mutation for linkage analysis to construct haplotypes containing mutations. (I) It clearly indicates whether the embryo is aneuploidy. Through the analysis of H and I, we can tell which embryo can be transferred (see J). (K) It is the process by which we transfer embryos into the uterus. This flowchart uses case 1 as an example
Genetic variation detection
Learn basic family information through genetic counseling. The hereditary cancer-related mutation was verified in family members by Sanger sequencing. Next, genomic DNA was extracted from the peripheral blood of couples and references, which was performed for library construction, probe capture, and karyomapping. The pathogenic criteria of the detected mutations were evaluated according to ACMG [24].
Embryo biopsy, single-cell WGA
The blastocyst (day 5 or day 6) is the most commonly used stage of embryo biopsy by far [25], while trophoblast culture is the most commonly used biopsied tissue [26, 27]. Due to the limited number of biopsied cells, we adopted multiple displacement amplification (MDA) for single-cell whole-genome amplification (WGA).
Karyomapping array, haplotype analysis, and CNV detection
Karyomapping is based on SNP linkage analysis flanking the mutation. We selected SNPs within 2M of upstream and downstream of the related mutation. According to the relevant guidance provided by ESHRE (European Society of Human Reproduction and Embryology), we should include a minimum of three informative SNP loci within this range [28]. This region length minimizes the impact of chromosomal recombination on linkage analysis. The reference is not only a member of the pedigree but also a crucial component for constructing haplotypes by linkage analysis. The SNPs are designed on the main principle that carriers are heterozygous and normal people are homozygous in couples. Besides, if the reference is the parent of the couple and carries the mutation, the selection of the reference SNP allele is required to be homozygous. If the reference is the offspring of the couple, there is no specific requirement for the selection of the reference SNP allele.
SNP information is obtained through karyomapping technology. After the SNPs were designed, haplotype analysis can be further verified. We construct haplotypes by couples and references in the pedigree. Based on available SNP locus information, haplotype analysis can determine whether the embryo carries the related mutation.
It can also detect copy number variations (CNVs) and their parental origin, including aneuploidy and partial deletion of chromosomes. And whether the embryo is aneuploid or not can be determined by visualizing the log R ratio (LRR) and B-allele frequency (BAF). LRR and BAF were used to estimate the CNVs. Because LRR is the logged ratio of observed probe intensity to expected intensity, any deviations from zero in this metric are evidence for copy number change. BAF is the proportion of the hybridized sample that carries the B allele as designated. In a normal sample, discrete BAFs of 0.0, 0.5, and 1.0 are expected for each locus (representing AA, AB, and BB).
Frozen embryo transfer (FET) and follow-up
The blastocysts with normal copy numbers that do not carry familial cancer-predisposing mutations were transferred into the uterus during frozen embryo transfer (FET) cycles. Amniotic fluid genetic testing was performed at 18–22 gestational weeks to validate the result of preimplantation genetic testing for monogenic disorders (PGT-M).
Results
Characteristics of included couples
A total of six families (7 cycles) were included in this study, all of which are inherited cancer-related gene families; 43 embryos from 7 PGT-M cycles were biopsied from the six couples. An average of 6.14 blastocysts were biopsied per cycle. Single-cell WGA was performed after obtaining TE cells, and the success rate of WGA was 100% (43/43). According to the ACMG, case 1, case 2, and case 6 were classified to pathogenic genes, and case 3 and case 4 were classified to likely pathogenic genes.
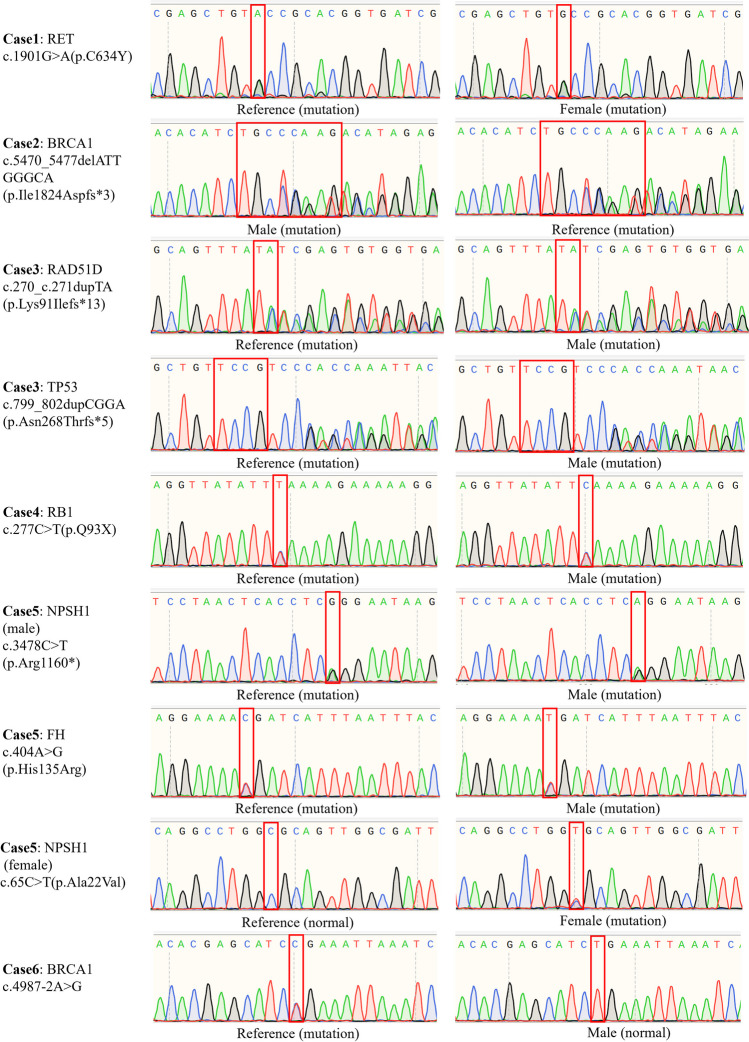
Case 1 is a familial medullary thyroid carcinoma (fMTC) that has been inherited within three generations (proband, mother, daughter). Both the proband and her mother have been diagnosed with medullary thyroid carcinoma. Case 2 was found to carry the BRCA1 gene because of his high myopia with his father. Therefore, the gene has a specific pathogenicity of breast cancer, so special attention should be paid to the vertical transmission of BRCA1 in the female offspring. In case 3, the proband came to the clinic with astrocytoma and was found to carry RAD51D and TP53 mutations, and it was found that the mutation originated from his father. These mutations are an inherited tumor susceptibility gene. The proband in case 4 was diagnosed with retinoblastoma, and the related gene was de novo RB1 mutation. This mutation has been passed on to offspring by amniotic fluid testing. In case 5, the proband developed renal cell carcinoma, and the FH mutation came from his father and was likely to be passed on to other offspring. In case 6, the reference was diagnosed due to delayed myelination in children with developmental delay, and trio whole exome sequencing (Trio WES) detected that his father carried the BRCA1 gene. Given the specific pathogenicity of BRCA1 and the fact that obtaining this mutation in female offspring greatly increases the risk of breast cancer [29], PGT-M is carried out in this couple to block the genetic tumor risk of offspring. The relevant risk genes and cycles of the included families can be found in Table 1, and the family trees show the inheritance of hereditary tumor susceptibility genes in detail (Supplementary Fig. 1). In addition, the Sanger sequencing results of susceptibility genes carried by various families, along with the corresponding primers, are presented in Fig. 2.
Fig. 2.
The results of gene mutation detection by Sanger sequencing of families
Strategy analysis
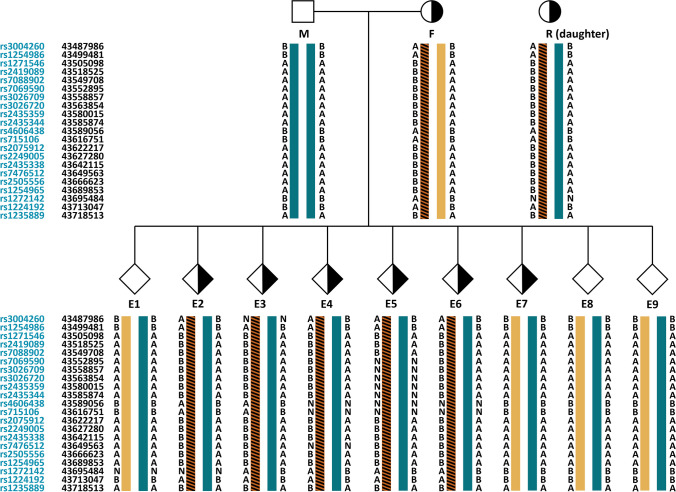
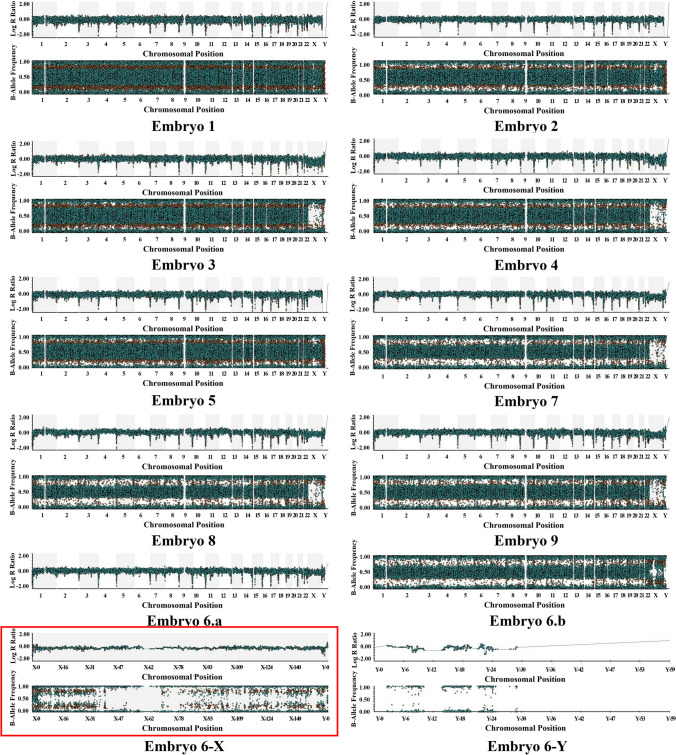
Taking case 1 as an example, the proband was the wife of the couple who had been diagnosed with medullary thyroid carcinoma when she came to our center. The daughter of the proband has been detected to carry the RET mutation. We can obtain SNP information for analysis using the karyomapping technique and select SNPs within 2M upstream and downstream of RET mutations for linkage analysis. The principle of informative SNP site selection is that the carrier is heterozygous, and the normal one is homozygous in couples. SNP linkage analysis based on the reference can construct haplotypes. We identified the haplotypes carrying the pathogenic gene mutation, which can analyze the RET mutation status of the blastocysts biopsied. We can analyze E1, E7, E8, and E9 do not carry the RET mutant gene. (See Fig. 3 for details.) CNV testing was performed on 9 embryos in one cycle, and we found that E1, E2, E3, E4, E5, E7, E8, and E9 were all euploidy embryos (Fig. 4). Based on the haplotype analysis and CNV results, E1, E7, E8, and E9 can be transferred.
Fig. 3.
Haplotype analysis of embryos in case 1. M means the male. F means the female. R means references, and E means embryos. This figure shows in detail how case 1 constructed haplotypes containing mutations and performed linkage analysis. Firstly, SNPs within 2M upstream and downstream of the mutation were selected, and the female (F) in the couple was known to be a carrier and the male (M) was a non-carrier. Therefore, the male SNP selects homozygous alleles, and the female SNP selects heterozygous alleles. Because their daughter (R) is a carrier, haplotypes containing mutations can be constructed according to reference (R) and then extrapolated to the mutation carrier of each embryo.
 : Male without hereditary cancer-related mutation.
: Male without hereditary cancer-related mutation.
 : Female with hereditary cancer-related mutation.
: Female with hereditary cancer-related mutation.
 : unknown gender without hereditary cancer-related mutation.
: unknown gender without hereditary cancer-related mutation.
 : unknown gender with hereditary cancer-related mutation
: unknown gender with hereditary cancer-related mutation
Fig. 4.
The CNV results in case 1. Embryo 6-a is the result of the log R Ratio of CNVs in embryo 6. Embryo 6-b is the result of the B-allele frequency of CNVs in embryo 6. Embryo 6-X and Embryo 6-Y represent the result plots of X and Y chromosome LRR and BAF of embryo 6, respectively
SNP linkage analysis and CNV diagnosis
For other families, we analyzed and diagnosed according to the above strategies for SNP linkage analysis and CNV diagnosis.
After testing of the pathogenic gene in the family and single-cell WGA of the blastocyst, screening was performed for the couple and reference by karyomapping. The haplotype was constructed by SNP linkage analysis to determine the pathogenic gene carrier status of embryos in case 1, case 2, case 3, case 4, case 5, and case 6 (Supplementary Tables 1, 2, 3, 4, 5, 6). CNV testing was then performed, and we can obtain embryonic aneuploidy information through visualizing LRR and BAF.
The results of haplotype analysis showed that the proportion of embryos without hereditary cancer-related mutations was 46.5%, and the proportion of embryos carrying hereditary cancer-related mutations reached 53.5%. The results of CNV showed that the proportion of euploid embryos reached 48.8%, and the proportion of chromosomally abnormal embryos reached 51.2%. The number of informative SNP alleles near the related mutation and the CNV results in case 1, case 2, case 3, case 4, case 5, and case 6 are presented in Table 2. Based on the results of haplotype linkage analysis and CNVs, it was comprehensively judged whether embryos could be transferred. A total of 15 transferable embryos were obtained, with an average of 2.14 transferable embryos per cycle. Of the 7 cycles, only cycle 1 in case 5 had no transferable embryos. Embryos are transferable in all other cycles.
Table 2.
The results of haplotype analysis and CNVs of embryos
| Couple | Cycle | Embryo | Gene | Mutation carrier status | Informative SNPs (2M) | CNV† | Outcomes |
|---|---|---|---|---|---|---|---|
| Case 1 | 1 | 1 | RET | Normal | 47 | 46,XN | Live birth |
| 1 | 2 | RET | Carrier | 49 | 46,XN | Abandoned | |
| 1 | 3 | RET | Carrier | 46 | 46,XN | Abandoned | |
| 1 | 4 | RET | Carrier | 46 | 46,XN | Abandoned | |
| 1 | 5 | RET | Carrier | 41 | 46,XN | Abandoned | |
| 1 | 6 | RET | Carrier | 47 | 46,XY(mosic80%)/47,XXY(mosic20%) | Abandoned | |
| 1 | 7 | RET | Normal | 50 | 46,XN | Not transfer | |
| 1 | 8 | RET | Normal | 49 | 46,XN | Not transfer | |
| 1 | 9 | RET | Normal | 49 | 46,XN | Not transfer | |
| Case 2 | 1 | 1 | BRCA1 | Normal | 24 | 46,XN | Live birth |
| 1 | 2 | BRCA1 | Normal | 23 | 46,XN | Not transfer | |
| 1 | 3 | BRCA1 | Normal | 23 | 46,XN | Not transfer | |
| 1 | 4 | BRCA1 | Normal | 17 | 46,XN | Not transfer | |
| 1 | 5 | BRCA1 | Carrier | 25 | 38,XN,-3,-7,-8,-12,-13,-15,-16,-17 | Abandoned | |
| 1 | 6 | BRCA1 | Carrier | 24 | 46,XN | Abandoned | |
| 1 | 7 | BRCA1 | Carrier | 24 | 51,XN,del(6)(q12-q27),+2,+7,+14,+15,+19 | Abandoned | |
| Case 3 | 1 | 1 | RAD51D; TP53 | Normal; Normal | 57; 33 | 46,XN | Live birth |
| 1 | 2 | RAD51D; TP53 | Normal; Normal | 64; 32 | 46,XN,dup(12)(q23.3-qter) | Abandoned | |
| 1 | 3 | RAD51D; TP53 | Carrier; carrier | 67; 34 | 46,XN,7p+ | Abandoned | |
| 1 | 4 | RAD51D; TP53 | Carrier; normal | 65; 34 | 52,XN,+1,+5,+6,+7,+21,+22 | Abandoned | |
| 1 | 5 | RAD51D; TP53 | Normal; Normal | 68; 35 | 46,XN | Not transfer | |
| 1 | 6 | RAD51D; TP53 | Normal; Normal | 62; 34 | 46,XN | Not transfer | |
| 1 | 7 | RAD51D; TP53 | Carrier; normal | 66; 34 | 47,XN,del(2)(q32.1-q37.3),+7 | Abandoned | |
| Case 4 | 1 | 1 | RB1 | Normal | 31 | 46,XN | Abortion |
| 1 | 2 | RB1 | Carrier | 30 | 46,XN,del(1)(pter-p31.3),3q- | Abandoned | |
| 1 | 3 | RB1 | Normal | 35 | 46,XN | Pregnancy | |
| Case 5 | 1 | 1 | FH; NPHS1‡ | Carrier; normal | 40; 22 | 46,XN,del(3)(q11.2-q29) | Abandoned |
| 1 | 2 | FH; NPHS1‡ | Carrier; normal | 32; 20 | 45,XN,-21 | Abandoned | |
| 1 | 3 | FH; NPHS1‡ | Carrier; carrier | 35; 24 | 45,XN,-22 | Abandoned | |
| 1 | 4 | FH; NPHS1‡ | Carrier; normal | 39; 23 | 47,XN,+8 | Abandoned | |
| 2 | 1 | FH; NPHS1‡ | Carrier; normal | 38; 23 | 47,XN,+21 | Abandoned | |
| 2 | 2 | FH; NPHS1‡ | Normal; Normal | 32; 20 | 46,XN | Pregnancy | |
| 2 | 3 | FH; NPHS1‡ | Carrier; carrier | 38; 23 | 45,XN,-22 | Abandoned | |
| 2 | 4 | FH; NPHS1‡ | Carrier; normal | 39; 22 | 47,XN,+4,-21,+22 | Abandoned | |
| 2 | 5 | FH; NPHS1‡ | Carrier; carrier | 39; 24 | 46,XN | Abandoned | |
| 2 | 6 | FH; NPHS1‡ | Carrier; carrier | 40; 24 | 47,XN,+16 | Abandoned | |
| 2 | 7 | FH; NPHS1‡ | Normal; carrier | 39; 24 | 46,XN | Abandoned | |
| 2 | 8 | FH; NPHS1‡ | NA; carrier | 39; 24 | 47,XN,+8 | Abandoned | |
| Case 6 | 1 | 1 | BRCA1 | Normal | 58 | 46,XN,upd(4)(q32.3-q34.3) | Pregnancy |
| 1 | 2 | BRCA2 | Normal | 57 | 45,XN,-21 | Abandoned | |
| 1 | 3 | BRCA3 | Carrier | 65 | 45,XN,-19 | Abandoned | |
| 1 | 4 | BRCA4 | Normal | 60 | 47,XN,+3 | Abandoned | |
| 1 | 5 | BRCA5 | Normal | 60 | 45,XN,upd(4)(q32.3-q34.3),-22 | Abandoned |
†Copy number variants
‡Indicates the number of informative SNPs within 1M upstream and downstream of the NPHS1 mutation
Frozen embryo transfer (FET) and follow-up
We selected embryos without hereditary cancer-related mutation for transfer into the uterus. Three of the six couples gave birth to healthy babies, and three are pregnant. Case 1, case 2, and case 3 all gave birth to healthy infants; none of the babies were found to carry the associated inherited mutation by amniotic fluid testing. In case 4, the couple transferred an embryo to obtain a clinical pregnancy, and amniotic fluid testing confirmed that the embryo did not carry the hereditary cancer-predisposing mutation, while the couple in case 6 has not tested yet due to insufficient pregnancy weeks. In case 5, embryos without hereditary cancer-predisposing mutation were successfully transplanted and obtained clinical pregnancy. But, the couple induced labor due to fetal megacystis during pregnancy.
Discussion
Cancer genetics is increasingly becoming integrated into the practice of modern medical oncology. Several tumors involved in this study are common familial hereditary tumors with clear family history. Individuals with inherited cancer-related mutations have a much higher risk of developing associated cancers than non-carriers. Taking the proto-oncogene BRCA1 as an example, both case 2 and case 6 carried the BRCA1 gene in the family. And study has shown that BRCA1 mutations account for 30–35% of all hereditary breast cancers overall [30]. The lifetime risk of developing breast cancer for these women ranges from 47 to 66% in BRCA1 mutation carriers, respectively [29, 31, 32].
Currently, both the psychological and financial costs of prevention and treatment for hereditary cancers are substantial, whether before or after birth. The clustering of hereditary cancer cases within families imposes a heavy burden on individuals, families, and healthcare systems [33–35].
We tried to use PGT-M to advance the prevention of hereditary tumors to primary prevention and to block the transmission of some cancer-associated mutations with specific pathogenicity or clear family history to offspring before pregnancy. At present, the centers mainly perform PGT-M for single-gene genetic diseases [36, 37].
Our center has been successfully applied to 6 families, all of which have obtained embryos without inherited cancer-related mutations. A total of 6 inherited cancer-related genes were involved in the six couples. These couples have all achieved clinical pregnancies, and 5 of these cases have been confirmed by amniotic fluid testing to be fetuses without the associated mutation. Our study provides feasible strategies for the prevention of hereditary tumors and has proven its effectiveness.
This research strategy has its obvious advantages. First, from the perspective of primary prevention, the transmission of cancer-related mutations with a clear family history to offspring is blocked, reducing the physical harm to pregnant women and the economic loss of the family. Second, in order to reduce the risk of allele dropout (ADO) during single-cell whole-genome amplification (WGA) [38], we avoid the effects of ADO according to haplotype analysis by karyomapping [39]. Because inherited tumors mostly exhibit an autosomally dominant pattern of inheritance [40], ADO in single-cell WGA will have serious consequences. Our research provides feasible technical support for PGT-M in the further improvement of cancer-related gene screening.
At the same time, our study had some limitations. First, the sensitivity of karyomapping in detecting CNVs is not as capable than that of next-generation sequencing (NGS) when it comes to detecting mosaic and small fragment deletions and duplications. Then, the implementation of our strategy requires complete family information, which places demands on the integrity of the family information. Finally, mutation identification and haplotype linkage analysis take more time and cost.
The use of PGT-M in hereditary tumors may be controversial due to racial, religious, or other ethical issues [41], and because of the diversity of cancer causes [42], PGT-M is not a guarantee of a healthy future. However, it is undeniable that in patients with a family history of hereditary tumors, blocking the susceptibility genes of inherited tumors can greatly reduce the probability of developing hereditary cancer, especially in the family with a specific pathogenic mutation or a strong family history, which has great positive significance for the individual, family, and society.
Our research demonstrated the feasibility of our strategy. In order to better serve families affected by hereditary tumors, deeper investigation and understanding of the mechanism behind hereditary cancer-related mutations are necessary in the future.
Supplementary information
SUPPLEMENTAL Figure 1. Family trees of all family were included. (TIF 1252 kb)
SUPPLEMENTAL TABLE 1. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the RET gene. (DOCX 30 kb)
SUPPLEMENTAL TABLE 2. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 21 kb)
SUPPLEMENTAL TABLE 3A. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the RAD51D gene. SUPPLEMENTAL TABLE 3B. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the TP53 gene. (DOCX 39 kb)
SUPPLEMENTAL TABLE 4. The results of haplotype linkage analysis of case 4 by SNPs within 2M of upstream and downstream of the RB1 gene. (DOCX 21 kb)
SUPPLEMENTAL TABLE 5A. The results of haplotype linkage analysis of case 5 by SNPs within 2M of upstream and downstream of the FH gene. SUPPLEMENTAL TABLE 5B. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from paternal chromosome. SUPPLEMENTAL TABLE 5C. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from maternal chromosome. (DOCX 38 kb)
SUPPMENTAL TABLE 6. The results of haplotype linkage analysis of case 6 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 28 kb)
Acknowledgements
We would like to thank all the participants that took part in the study. This work was supported by the National Natural Science Foundation of China for the National Key R&D Program of China (2019YFA0110900) to Yingpu Sun.
Author contribution
Chuanju Chen: conceptualization (supporting), data curation (lead), formal analysis (equal), validation (equal), visualization (lead), writing – original draft preparation (lead), writing – review and editing. Hao Shi: conceptualization (lead), formal analysis (equal), investigation (lead), methodology (lead), project administration (lead), supervision, writing – review and editing (lead). Wenbin Niu: formal analysis (equal), resources (equal). Xiao Bao: formal analysis (equal), resources (equal). Jingya Yang: validation (equal). Haixia Jin: formal analysis (equal), resources (equal). Wenyan Song: formal analysis (equal), resources (equal). Yingpu Sun: funding acquisition (lead), supervision (lead).
Funding
This work was supported by funding from the National Natural Science Foundation of China for the National Key R&D Program of China (2019YFA0110900) to Y.-P.S.
Data availability
The data that support the results of this study can be obtained from the corresponding author upon reasonable request.
Declarations
Ethics approval
Ethical approval to conduct this retrospective study was obtained from the Internal Review Board of The First Affiliated Hospital of Zhengzhou University (Ethic no. 2023-KY-0361).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chuanju Chen and Hao Shi contributed equally to this work.
References
- 1.Siegel RL, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Fitzmaurice C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncol. 2018;4(11):1553–1568. doi: 10.1001/jamaoncol.2018.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23(38):6445–6470. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 4.Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23(2):276–292. doi: 10.1200/JCO.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 6.Fillon M. Breast cancer recurrence risk can remain for 10 to 32 years. CA Cancer J Clin. 2022;72(3):197–199. doi: 10.3322/caac.21724. [DOI] [PubMed] [Google Scholar]
- 7.Sakorafas GH, Tsiotou AG. Genetic predisposition to breast cancer: a surgical perspective. Br J Surg. 2000;87(2):149–162. doi: 10.1046/j.1365-2168.2000.01347.x. [DOI] [PubMed] [Google Scholar]
- 8.Handyside AH, et al. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344(6268):768–770. doi: 10.1038/344768a0. [DOI] [PubMed] [Google Scholar]
- 9.Bickerstaff H, et al. Clinical application of preimplantation genetic diagnosis. Hum Fertil (Camb). 2001;4(1):24–30. doi: 10.1080/1464727012000199221. [DOI] [PubMed] [Google Scholar]
- 10.Treff NR, et al. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertil Steril. 2013;99(5):1377–1384.e6. doi: 10.1016/j.fertnstert.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Qubbaj W, et al. First successful application of preimplantation genetic diagnosis and haplotyping for congenital hyperinsulinism. Reprod Biomed Online. 2011;22(1):72–79. doi: 10.1016/j.rbmo.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, et al. A strategy using SNP linkage analysis for monogenic diseases PGD combined with HLA typing. Clin Genet. 2020;98(2):138–146. doi: 10.1111/cge.13770. [DOI] [PubMed] [Google Scholar]
- 13.Yan L, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112(52):15964–15969. doi: 10.1073/pnas.1523297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thornhill AR, et al. Karyomapping – a comprehensive means of simultaneous monogenic and cytogenetic PGD: comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–356. doi: 10.1007/s10815-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harton GL, et al. ESHRE PGD consortium best practice guidelines for fluorescence in situ hybridization-based PGD. Hum Reprod. 2011;26(1):25–32. doi: 10.1093/humrep/deq230. [DOI] [PubMed] [Google Scholar]
- 16.Ricciardiello L, Ahnen DJ, Lynch PM. Chemoprevention of hereditary colon cancers: time for new strategies. Nat Rev Gastroenterol Hepatol. 2016;13(6):352–361. doi: 10.1038/nrgastro.2016.56. [DOI] [PubMed] [Google Scholar]
- 17.Grunfeld E, et al. Family caregiver burden: results of a longitudinal study of breast cancer patients and their principal caregivers. Cmaj. 2004;170(12):1795–1801. doi: 10.1503/cmaj.1031205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishanina E, et al. Use of labour induction and risk of cesarean delivery: a systematic review and meta-analysis. Cmaj. 2014;186(9):665–673. doi: 10.1503/cmaj.130925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, et al. Preimplantation genetic diagnosis using combined strategies on a breast cancer patient with a novel genomic deletion in BRCA2. J Assist Reprod Genet. 2014;31(12):1719–1726. doi: 10.1007/s10815-014-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasper MJ, Liebelt J, Hussey ND. Preimplantation genetic diagnosis for BRCA1 exon 13 duplication mutation using linked polymorphic markers resulting in a live birth. Prenat Diagn. 2008;28(4):292–298. doi: 10.1002/pd.1925. [DOI] [PubMed] [Google Scholar]
- 21.Rechitsky S, et al. Preimplantation genetic diagnosis for cancer predisposition. Reprod Biomed Online. 2002;5(2):148–155. doi: 10.1016/s1472-6483(10)61617-3. [DOI] [PubMed] [Google Scholar]
- 22.Girardet A, et al. First preimplantation genetic diagnosis of hereditary retinoblastoma using informative microsatellite markers. Mol Hum Reprod. 2003;9(2):111–116. doi: 10.1093/molehr/gag014. [DOI] [PubMed] [Google Scholar]
- 23.Shenfield F, et al. Taskforce 5: preimplantation genetic diagnosis. Hum Reprod. 2003;18(3):649–651. doi: 10.1093/humrep/deg110. [DOI] [PubMed] [Google Scholar]
- 24.Biesecker LG, Harrison SM. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med. 2018;20(12):1687–1688. doi: 10.1038/gim.2018.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimadomo D, et al. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Hum Reprod Update. 2020;26(4):453–473. doi: 10.1093/humupd/dmaa019. [DOI] [PubMed] [Google Scholar]
- 26.Kokkali G, et al. Birth of a healthy infant following trophectoderm biopsy from blastocysts for PGD of beta-thalassaemia major. Hum Reprod. 2005;20(7):1855–1859. doi: 10.1093/humrep/deh893. [DOI] [PubMed] [Google Scholar]
- 27.McArthur SJ, et al. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005;84(6):1628–1636. doi: 10.1016/j.fertnstert.2005.05.063. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho F, et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020(3):hoaa018. doi: 10.1093/hropen/hoaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee A, Moon BI, Kim TH. BRCA1/BRCA2 Pathogenic variant breast cancer: treatment and prevention strategies. Ann Lab Med. 2020;40(2):114–121. doi: 10.3343/alm.2020.40.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, et al. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J Clin Oncol. 2006;24(6):863–871. doi: 10.1200/JCO.2005.03.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narod SA. Which genes for hereditary breast cancer? N Engl J Med. 2021;384(5):471–473. doi: 10.1056/NEJMe2035083. [DOI] [PubMed] [Google Scholar]
- 33.Carrera PM, Kantarjian HM, Blinder VS. The financial burden and distress of patients with cancer: understanding and stepping-up action on the financial toxicity of cancer treatment. CA Cancer J Clin. 2018;68(2):153–165. doi: 10.3322/caac.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradley CJ. Cancer, financial burden, and medicare beneficiaries. J Clin Oncol. 2017;35(22):2461–2462. doi: 10.1200/JCO.2017.73.1877. [DOI] [PubMed] [Google Scholar]
- 35.Li N, et al. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the Global Burden of Disease Study 2017. J Hematol Oncol. 2019;12(1):140. doi: 10.1186/s13045-019-0828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chada AR, et al. Trends and outcomes for preimplantation genetic testing for monogenic disorders in the United States, 2014-2018. Fertil Steril. 2022;118(6):1190–1193. doi: 10.1016/j.fertnstert.2022.08.854. [DOI] [PubMed] [Google Scholar]
- 37.Lee I, et al. Utilization of preimplantation genetic testing for monogenic disorders. Fertil Steril. 2020;114(4):854–860. doi: 10.1016/j.fertnstert.2020.05.045. [DOI] [PubMed] [Google Scholar]
- 38.Piyamongkol W, et al. Detailed investigation of factors influencing amplification efficiency and allele drop-out in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003;9(7):411–420. doi: 10.1093/molehr/gag051. [DOI] [PubMed] [Google Scholar]
- 39.Handyside AH, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47(10):651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- 40.Sherr CJ. Principles of tumor suppression. Cell. 2004;116(2):235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 41.Ethics of preimplantation genetic diagnosis for cancer. Lancet Oncol. 2006;7(8):611. [DOI] [PubMed]
- 42.Luzzatto L, Pandolfi PP. Causality and chance in the development of cancer. N Engl J Med. 2015;373(1):84–88. doi: 10.1056/NEJMsb1502456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL Figure 1. Family trees of all family were included. (TIF 1252 kb)
SUPPLEMENTAL TABLE 1. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the RET gene. (DOCX 30 kb)
SUPPLEMENTAL TABLE 2. The results of haplotype linkage analysis of case 1 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 21 kb)
SUPPLEMENTAL TABLE 3A. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the RAD51D gene. SUPPLEMENTAL TABLE 3B. The results of haplotype linkage analysis of case 3 by SNPs within 2M of upstream and downstream of the TP53 gene. (DOCX 39 kb)
SUPPLEMENTAL TABLE 4. The results of haplotype linkage analysis of case 4 by SNPs within 2M of upstream and downstream of the RB1 gene. (DOCX 21 kb)
SUPPLEMENTAL TABLE 5A. The results of haplotype linkage analysis of case 5 by SNPs within 2M of upstream and downstream of the FH gene. SUPPLEMENTAL TABLE 5B. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from paternal chromosome. SUPPLEMENTAL TABLE 5C. The results of haplotype linkage analysis of case 5 by SNPs within 1M of upstream and downstream of the NPHS1 gene from maternal chromosome. (DOCX 38 kb)
SUPPMENTAL TABLE 6. The results of haplotype linkage analysis of case 6 by SNPs within 2M of upstream and downstream of the BRCA1 gene. (DOCX 28 kb)
Data Availability Statement
The data that support the results of this study can be obtained from the corresponding author upon reasonable request.