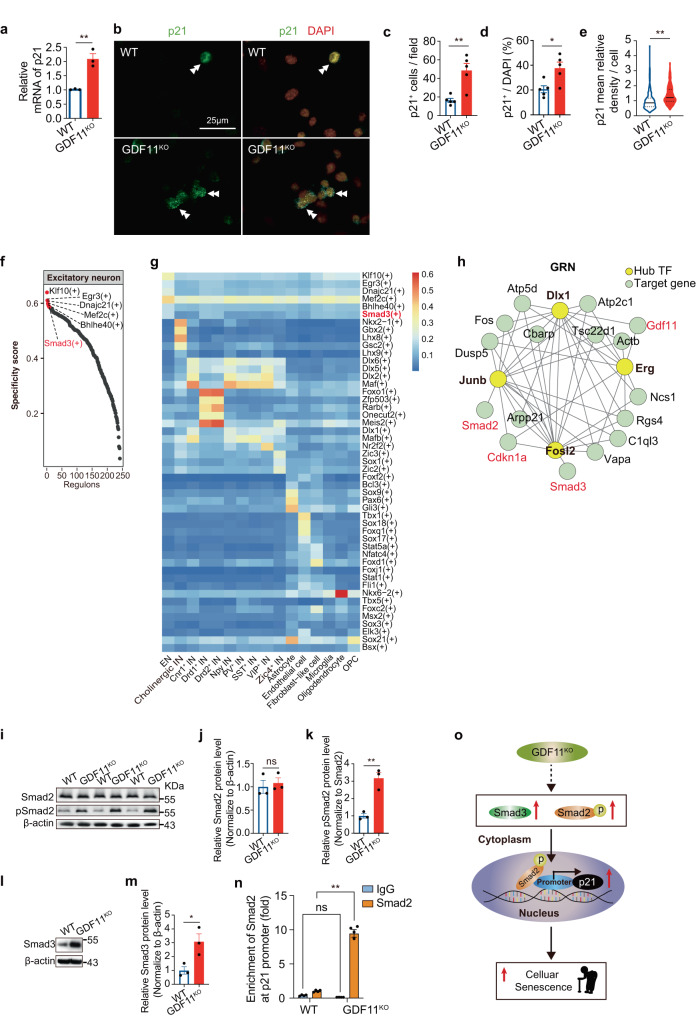
Fig. 8. Loss of GDF11 upregulates pSmad2 and Smad3 and enhances Smad2 binding to the promoter of p21.
a Quantification by qPCR of the relative p21 mRNA in the GDF11KO and WT Neuro-2a cells (n = 3 clones). b–e Immunofluorescence representative images (b) and quantification of the number of p21+ cells per field (c, n = 5 fields/group), the proportion of p21+ cells (d, n = 6 fields/group) or the average gray value of p21 per cell (e, GDF11KO, n = 420 cells; WT, n = 280 cells) in the GDF11KO and WT Neuro-2a cells. Scale bar, 25 μm. Examples of the p21+ cells are indicated by double arrowheads. f–h The same snRNA-seq data were used, as described in Fig. 5a. f Rank for regulons in the EN based on regulon specificity score (RSS). The EN were obtained from the Cg2 of the “KO” mice and the “Ctrl” mice aged 4–5 M. g Regulons activity analysis based on area under the curve (AUC) in the identified cell types in snRNA-seq of the “KO” mice and the “Ctrl” mice aged 4–5 M. The activity of regulon Smad3 (highlighted in red) is high in the EN. h Cytoscape network visualization of genes including GDF11, Cdkn1a (p21), Smad2, Smad3 (highlighted in red) and their transcription factors (TFs, yellow). i–m Representative images (i and l) and quantification by densitometry of western blot analysis of Smad2 (j), phosphorylated Smad2 (pSmad2, k) and Smad3 (m) in the total protein extracted from the GDF11KO and WT Neuro-2a cells (n = 3 biological repeats/group). n ChIP-qPCR assessment of the enrichment of Smad2 at the promoter of Cdkn1a/p21 in the GDF11KO and WT Neuro-2a cells (n = 3 biological repeats/group). o A proposed working model for loss of GDF11 on cellular senescence. Loss of GDF11 upregulates pSmad2, enhances nuclear entry of Smad2/3 tricomplex and then Smad2 binds to the promoter of p21 and promotes the pro-senescence factor p21 transcription, and eventually causes cellular senescence. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01 and “ns” indicates not significant. a (P = 0.0037), c (P = 0.0033), d (P = 0.0157), e (P < 0.0001), j (P = 0.6648), k (P = 0.0040) and m (P = 0.0299), unpaired two-tailed t test. n (IgG: WT versus GDF11KO, P = 0.57; Smad2: WT versus GDF11KO, P < 0.001), two-way ANOVA with Sidak’s test. Source data are provided with this paper.