Fig. 1.
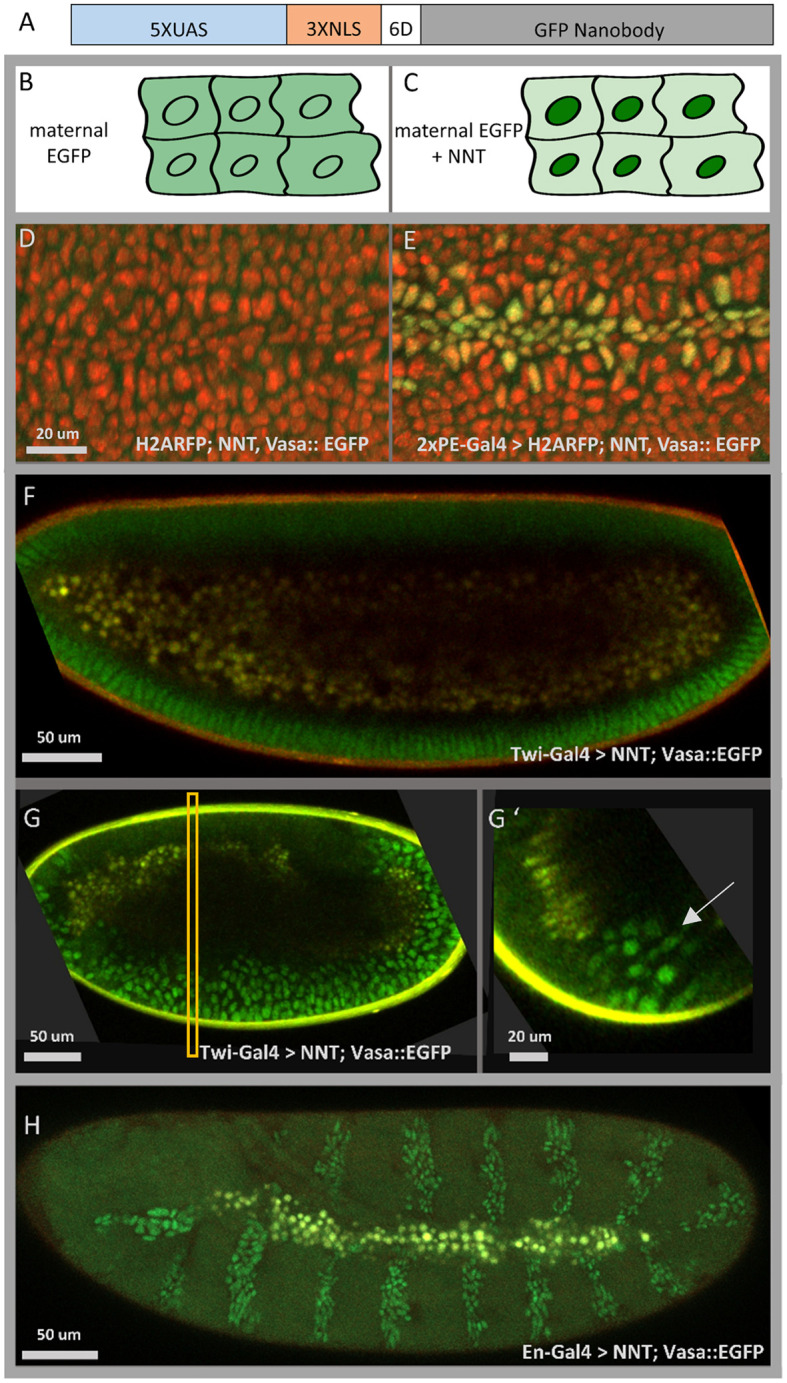
The NaNuTrap (NNT) method. (A) The 5×UAS::NNT construct: a GFP nanobody is fused to three nuclear localization sequences (3×NLS) with a six aspartate (6D) linker sequence and expression driven by the Gal4/UAS system (5×UAS). (B,C) Schematic illustrating the NNT method. (B) Maternally deposited GFP is distributed evenly in the embryo. (C) When the nanobody construct NNT is also expressed, it drives the already matured GFP into the cell nuclei. (D,E) Microscope stills from live in vivo imaging sessions in which the maternal driver Vasa is used to deposit GFP (Vasa::EGFP, green) in a His2Av-mRFP1 labelled (red) embryo (stage 10) (D); however, nuclear signal from GFP (green) is observed only when co-expressed with the NNT construct (E). (F) In the presence of maternally deposited GFP, the NNT construct, when expressed zygotically in the mesoderm and midline using the Twi-Gal4 driver, can localize GFP into the nuclei of the presumptive mesoderm cells in the cellular blastoderm (F, stage 5). (G,G′) Two-photon images of the collapsing furrow (stage 7/8); embryo genotype is the same as in F. Longitudinal section (G) and cross-section (G′) of the same 3D reconstructed image. Yolk autofluorescence appears in yellow, while signal from GFP-expressing nuclei is green. The rectangle in G indicates the position of the cross-section view shown in G′. Arrow indicates mesoderm cell nuclei. (H) Microscope still from a live in vivo imaging session (see Movie 4) of an embryo expressing NNT using the En-Gal4 driver; EGFP was deposited maternally.
