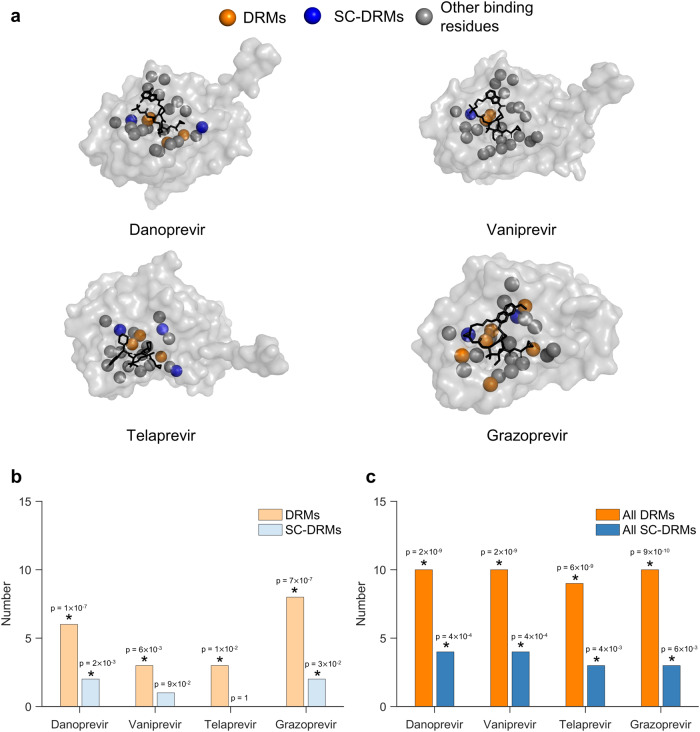
Fig. 5. SC-DRMs appear to impact binding of NS3 drugs through direct interactions.
a Binding residues of drugs shown on the crystal structure of the NS3 protein-drug complexes (PDB ID: 3M5L [10.2210/pdb3m5l/pdb] for danoprevir, 3SU3 [10.2210/pdb3su3/pdb] for vaniprevir, and 3SV6 [10.2210/pdb3sv6/pdb] for telaprevir and 3SUD [10.2210/pdb3sud/pdb] for grazoprevir). The carbon-alpha atoms of the identified drug-binding residues are shown in colored spheres. The drug-binding residues associated with DRMs and SC-DRMs for each drug are shown in green and blue, respectively, while those that do no fall under DRMs are shown in gray. Drugs in each structure are shown as black sticks. The NS3 residues within 5 Å7D2of drug atoms were considered as drug-binding residues. b, c Statistical significance of the number of (b) drug-specific DRMs/SC-DRMs and (c) all DRMs/SC-DRMs in binding residues of each of the four considered drugs. Here, drug-specific DRMs/SC-DRMs are listed in Table 1 for each of the four drugs, while all DRMs/SC-DRMs refer to the DRMs/SC-DRMs known for all drugs. The p-value measures the probability of observing by a random chance at least the observed number of DRMs or SC-DRMs among all binding residues for each drug (one-sided test; see “Methods” section for details). Results with p-value < 0.05 are marked with a star on the top of each bar. Source data and exact p-values are provided as a Source Data file.