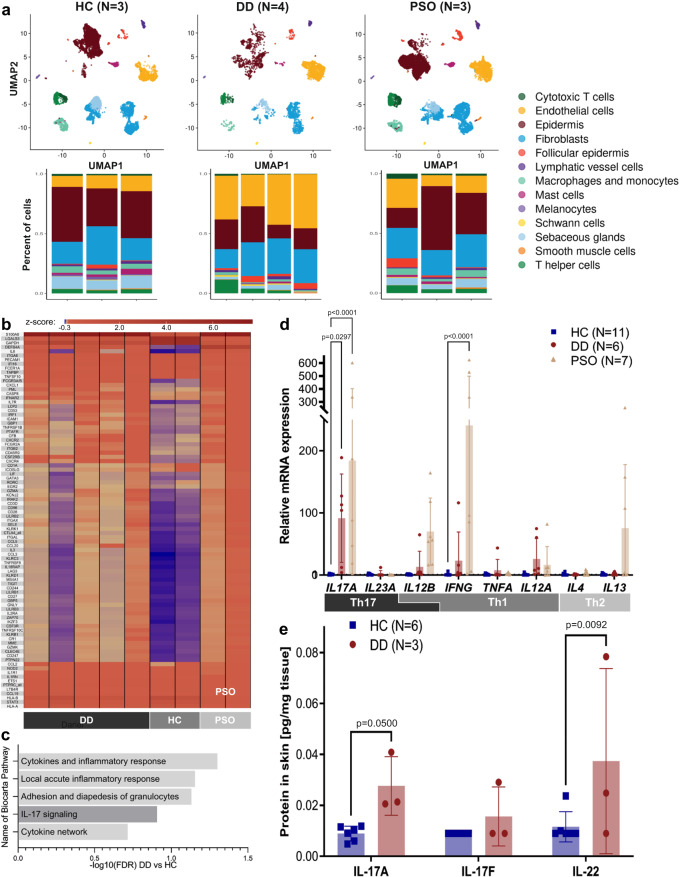
Fig. 1. Enhanced expression of Th17-related genes and cytokines in the skin of patients with Darier disease (DD).
a UMAP analysis (upper panel) and percentage of different cell types (lower panel) determined by scRNA-seq analysis of skin samples from four DD patients (lesional skin, N = 4, 3746 cells). Results were compared to publicly available scRNA-seq data of psoriasis (PSO, lesional skin, N = 3, 11417 cells) and healthy controls (HC, N = 3, 12817 cells)17. b Heat map of differentially expressed (DE) genes expressed as Z-score (DD vs HC, fold change > 1.5) as determined by NanoString nCounter analysis using the Immunology_V2 panel in skin of untreated DD patients (N = 5), healthy controls (N = 2) and psoriasis patients (N = 2). c Biocarta pathways most enriched in DD patients as compared to HC, as determined by NanoString nCounter and Gene Set Enrichment Analysis. Pathways are ordered according to their normalized enrichment scores. Bars represent -log10 of False Discovery Rates (FDR) (See Suppl. Table 2 for complete table). d Relative mRNA expression (qRT-PCR) of Th1, Th2, and Th17-associated cytokines normalized to housekeeping gene ACTB and relative to HC. Bars represent means, error bars represent standard deviations. p-values were calculated using 2-way ANOVA with Dunnett multiple comparison correction (all against HC). e Protein expression of Th17-related cytokines in the skin of HC and DD patients as determined by bead-based immunoassays (LegendPlex analysis). p-values were calculated using 2-way ANOVA. Error bars represent standard deviation.