Abstract
The COPI coatomer subunit α-COP has been shown to co-precipitate mRNA in multiple settings, but it was unclear whether the interaction with mRNA was direct or mediated by interaction with an adapter protein. The COPI complex often interacts with proteins via C-terminal dilysine domains. A search for candidate RNA binding proteins with C-terminal dilysine motifs yielded Nucleolin, which terminates in a KKxKxx sequence. This protein was an especially intriguing candidate as it has been identified as an interacting partner for Survival Motor Neuron protein (SMN). Loss of SMN causes the neurodegenerative disease Spinal Muscular Atrophy. We have previously shown that SMN and α-COP interact and co-migrate in axons, and that overexpression of α-COP reduced phenotypic severity in cell culture and animal models of SMA. We show here that in an mRNA independent manner, endogenous Nucleolin co-precipitates endogenous α-COP and ε-COP but not β-COP which may reflect an interaction with the so-called B-subcomplex rather a complete COPI heptamer. The ability of Nucleolin to bind to α-COP requires the presence of the C-terminal KKxKxx domain of Nucleolin. Furthermore, we have generated a point mutant in the WD40 domain of α-COP which eliminates its ability to co-precipitate Nucleolin but does not interfere with precipitation of partners mediated by non-KKxKxx motifs such as the kainate receptor subunit 2. We propose that via interaction between the C-terminal dilysine motif of Nucleolin and the WD40 domain of α-COP, Nucleolin acts an adaptor to allow α-COP to interact with a population of mRNA.
Keywords: COPI coatomer, nucleolin, spinal muscular atrophy, mRNA transport, neurodegeneration
Introduction
Spinal Muscular Atrophy (SMA) is an autosomal recessive neurodegenerative disease caused by mutation in the SMN1 gene and characterized by loss of the alpha motor neurons (reviewed in [1]. The Survival Motor Neuron (SMN) protein is required for proper biogenesis and assembly of small nuclear ribonucleoproteins (snRNPs) [2,3] and is also required for proper splicing of mRNA [4–6]. Beyond its involvement in splicing machinery assembly and function, SMN has been shown to regulate translation [7,8] and in cultured neurons SMN has repeatedly been implicated in the transport of mRNA throughout the axon and growth cone to promote local translation [9–12]. Most recently, it was shown that in cultured motor neurons, loss of SMN impairs assembly of ribosomes in axon growth cones inhibiting the local translation response that is required to respond to growth guidance cues [13].
The α-COP protein encoded by the COPA gene is a member of the heptameric COPI coatomer complex, which functions primarily in Golgi-ER retrograde transport, endosomal trafficking and autophagy [14]. In neuronal cells, α-COP is found throughout the axon and in the growth cone where it binds and co-localizes with the SMN, an interaction mediated by exon 2b of SMN [15,16]. In cultured mouse neurons, we found that knockdown of Copa significantly reduced the growth of both dendrites and axons [17]. Transgenic expression of Copa in cell culture and animal models of SMA ameliorates disease phenotype, restoring neurite length in NSC-34 cells, normal motor neuron axon morphology in Zebrafish, and increasing survival in a mouse model of SMN [16–18].
COPI subunits have frequently been detected in the axonal proteome [19–21], which we hypothesize reflects a unique function of COPI in this highly specialized neuronal compartment. COPI subunits have been shown to bind mRNA, first in experiments showing that COPI binding to the 5′-UTR regulated translation of the asialoglyoprotein receptor (ASGR) [22], then by experiments in yeast demonstrating that COPI dysfunction resulted in mis-localization of mRNA [23,24], followed by the finding that β-COP bound and transported kappa-opioid receptor mRNA [25]. RNA immunoprecipitation (RIP) from the motor neuron-like NSC-34 cell line showed that α-COP bound specifically to mRNA, including numerous transcripts that had been reported in axonal transcriptomes [26].
We set out to identify potential RNA binding proteins (RBPs) as candidates to interact with COPI coatomer and act as a bridge, allowing COPI vesicles to transport mRNA. One such candidate RBP was Nucleolin. Nucleolin was identified early on as interacting with SMN [27,28] and in hTERT fibroblasts with low levels of SMN, the subcellular localization of Nucleolin is disturbed resulting in altered localization of mTOR mRNA [29]. Nucleolin also has a high affinity for transcripts containing G-quadruplexes, a motif which was enriched in the α-COP RIP dataset [30,31]. In cultured neurons, Nucleolin is detected in the axonal compartment [32], and has been shown to play a role in regulating axon growth by mediating local translation in axons [33–35].
We report here findings that Nucleolin co-immunoprecipitates α-COP in an RNA-independent manner. The interaction is mediated by the C-terminal dilysine motif of Nucleolin interacting with the WD40 domain of α-COP. α-COP can immunoprecipitate known Nucleolin interacting mRNA, and in cells with stable knockdown of Nucleolin, the abundance of known Nucleolin interacting transcripts was reduced in α-COP RIPs. An α-COP mutant that was unable to bind Nucleolin was less able to restore neurite outgrowth in a mouse motor neuron-like cell line. Taken together, these data identify Nucleolin as a novel α-COP binding partner and a candidate adapter bringing specific mRNA to the COPI complex to support axonal growth and maintenance in motor neurons, and this interaction is required to fully support neurite outgrowth.
Results
Endogenous α-COP co-immunoprecipitates with Nucleolin
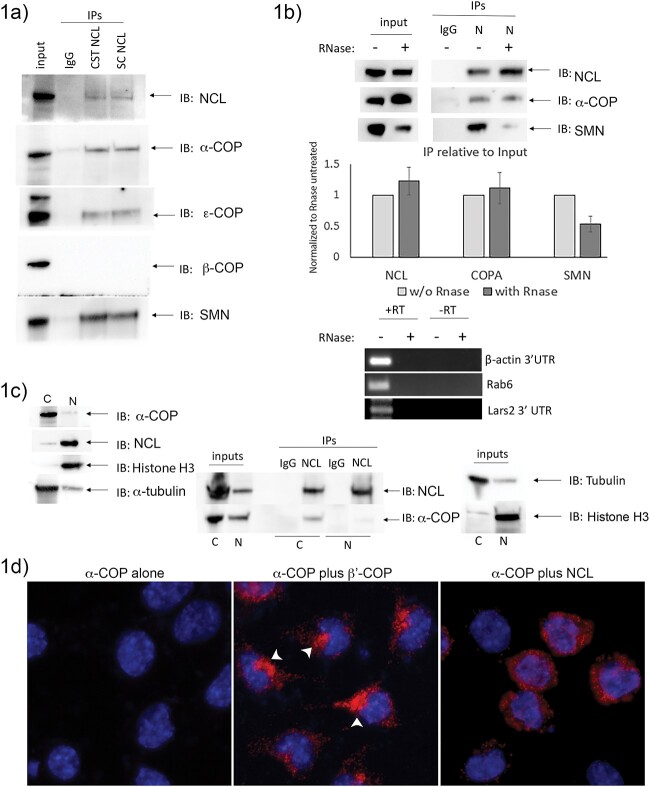
We began by confirming interaction between endogenous Nucleolin and endogenous α-COP. We carried out co-immunoprecipitation (co-IP) experiments from 293-TT whole cell lysates using either a rabbit polyclonal antibody (Cell Signaling, D4C70) or a mouse monoclonal (Santa Cruz, H-6) antibody against Nucleolin. Both antibodies efficiently and cleanly immunoprecipitated Nucleolin compared to IgG controls. Blotting back for COPI subunits showed that α-COP and ε-COP co-IP’d, but β-COP did not, indicating that the COPI/Nucleolin interaction may be limited to the B-subcomplex comprised of the α/β′/ε COPI subunits rather than an intact COPI heptamer [36]. This interaction was confirmed by immunoprecipitations from mouse brain as well as two murine cell lines (N2a and NSC-34) (not shown). Western blotting with an antibody specific to human SMN confirmed that SMN also co-IP’d, which was expected as this protein is known to co-precipitate with both Nucleolin and α-COP (Fig. 1a). Previously, co-IP between Nucleolin and SMN was shown to be RNAse sensitive, indicating that the binding was mediated by the presence of RNA [27]. To determine if the ability of Nucleolin to co-IP α-COP was similarly RNA dependent, we treated lysates with RNAse A prior to immunoprecipitation. As shown in Fig. 1b, approximately equal amounts of α-COP co-IP’d with Nucleolin in RNase treated samples compared to controls. Quantification of three replicate co-immunoprecipitation experiments by ImageJ densitometry demonstrates that the percent α-COP that co-IP’d with Nucleolin relative to the input was not significantly affected by RNAse treatment. We do observe reduced co-IP of SMN with Nucleolin in RNase treated samples (0.535 +/− 0.13-fold in RNAse treated IPs compared to untreated). The fact that the SMN co-IP was reduced rather than eliminated here may reflect the fact that this experiment was performed on whole cell lysate whereas the original was performed on nuclear extract. The RNase efficiently degraded the RNA as PCR for several transcripts were blank in treated samples (Fig. 1b, bottom panel).
Figure 1.
α-COP co-immunoprecipitates Nucleolin. (a) Representative Western blot showing immunoprecipitations from HEK-293TT whole cell lysate with either rabbit polyclonal (CST NCL) or mouse monoclonal (SC NCL) antibodies. Immunoblotting with antibody against Nucleolin demonstrates immunoprecipitation with both antibodies compared to IgG control (mixed mouse and rabbit IgG). Blotting with antibodies against COPI complex subunits shows co-immunoprecipitation of α-COP and ε-COP but not β-COP. (b) Whole cell HEK 293TT lysates were split and treated with RNAse A followed by immunoprecipitation of Nucleolin. A representative Western blot shows that in both control and RNAse treated samples, α-COP co-immunoprecipitates with Nucleolin. Successful degradation of RNA was confirmed by conversion to cDNA followed by PCR. Multiple primer sets demonstrate that signal is only present in the untreated samples and absent from the samples treated with RNase. (c) Representative Western blot from fractionated 293-TT cells showing the localization of Nucleolin and α-COP in the α-tubulin positive cytoplasm (C) and the Histone H3 positive nuclear fraction (N). Representative Western blot showing the results of Nucleolin immunoprecipitation from cytoplasmic (C) and nuclear (N) fractions. As expected, the Nucleolin primarily co-precipitates α-COP from the cytoplasm. (d) Proximity ligation assay was performed on fixed mouse neuroblastoma cells (N2A). Negative control slides (mouse monoclonal α-COP antibody alone) show clean PLA background (red). Positive control coverslips combined mouse monoclonal α-COP antibody and rabbit polyclonal β′-COP, resulting in a strong PLA signal with predominantly Golgi localization (white arrowheads) as expected from COPI coatomer localization. Finally, experimental coverslips combined the mouse monoclonal α-COP with rabbit polyclonal Nucleolin antibody, resulting in a clear cytoplasmic PLA signal. Nuclei are visualized with DAPI.
Although the bulk of Nucleolin protein resides in the nucleolus, it has long been appreciated that Nucleolin is able to shuttle between the nucleus and the cytoplasm where it impacts translational regulation and mRNA stability and transport [35,37–39]. Given that α-COP is predominantly localized to the Golgi apparatus, we assume that the interaction between Nucleolin and COPI subunits is a cytoplasmic event. To confirm that Nucleolin is present in the cytoplasm of 293-TT cells, we used a modified hypotonic lysis protocol and analyzed the resulting fractions by Western blot for Nucleolin and α-COP with α-tubulin to confirm the integrity of the cellular fractionation and Histone H3 to confirm the purity of the nuclear fraction. Nucleolin was found in the Histone H3 positive nuclear fraction, but detectable amounts were also present in the α-tubulin enriched cytoplasmic fraction, which contained the bulk of the α-COP (Fig. 1c). Co-immunoprecipitation of α-COP with Nucleolin was largely restricted to the cytoplasmic fraction despite detectable low levels of α-COP in the nuclear lysate. This confirms that sufficient cytoplasmic Nucleolin is present in 293-TT lysate to interact with cytoplasmic α-COP. To further evidence that α-COP binds to Nucleolin and the subcellular localization of this interaction, we used the proximity ligation assay (PLA) to detect these proteins in differentiated N2A cells (Fig. 1d). When performed with antibody to α-COP alone, there was no detectable red PLA signal. As a positive control, we performed the assay with antibodies to α-COP and β′-COP. As expected, this resulted in a robust PLA signal with predominantly Golgi-like distribution (white arrowheads). The PLA signal when using α-COP and Nucleolin antibodies was predominantly cytoplasmic, in agreement with the co-IP experiments performed on fractionated cell lysates. Comparable results were obtained on cultured NSC-34 cells (not shown). Taken together, these data confirm that the interaction between α-COP and Nucleolin is direct and occurs in the cytoplasm.
α-COP N-terminus interacts with Nucleolin C-terminus
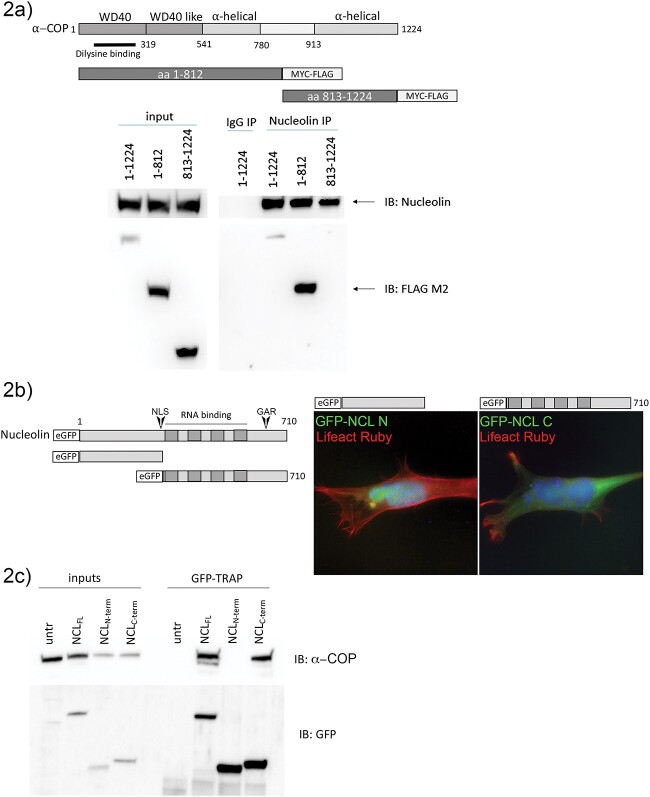
To further understand the biological basis of the interaction between α-COP and Nucleolin, we set out to determine which protein sequences were required. The α-COP protein contains an N-terminal WD40 domain, which mediates binding to traditional COPI vesicle cargo as well as binding to the β′-COP subunit [40]. The C-terminal portion of α-COP is more α-helical and unstructured and mediates the interaction with ε-COP [41]. We cloned N-terminal and C-terminal fragments of α-COP into a MYC-FLAG tagged expression vector, dividing the protein at amino acid 812. When Nucleolin was IP’d from 293-TT cells expressing either the full-length α-COP or the fragments, only the full-length and 1–812 region were detectable after blotting back with anti-FLAG antibody, indicating that the N-terminal portion of α-COP mediates its association with Nucleolin (Fig. 2a). In a complementary experiment, we subcloned GFP-tagged Nucleolin to produce an N-terminal fragment containing the intrinsically disordered domain and a C-terminal fragment containing the RNA recognitions motifs (RRMs) and the Glycine/Arginine rich region (GAR). The N-terminal region of Nucleolin can be heavily phosphorylated and mediates interaction with chromatin and also contains a bipartite nuclear localization signal (NLS) [42–44]. The C-terminal domain is required for Nucleolin interaction with and stabilization of G-quadruplex structures and mediates its subcellular localization in neurons [30,33]. Figure 2b shows a schematic of the N and C-terminal truncations as well as representative micrographs of NSC-34 cells co-transfected with LifeAct Ruby and truncated GFP-NCL showing that the N-terminal fragment containing the NLS does localize to the DAPI positive nucleus as expected. Using GFP-TRAP, we show that endogenous α-COP binds full-length GFP-Nucleolin and its GFP-tagged C-terminus but not the N-terminus. GFP-TRAP was performed on whole cell lysates in 1% NP40 to fully release both the nuclear and cytoplasmic compartments before co-immunoprecipitation was performed (Fig. 2c). Taken together, these experiments clearly demonstrate that the interaction between Nucleolin and α-COP requires the N-terminus of α-COP and the C-terminus of Nucleolin.
Figure 2.
The N-terminus of α-COP interacts with the C-terminus of Nucleolin. (a) Schematic of α-COP protein structure and Myc-FLAG tagged truncation mutants. A representative Western blot from HEK 293TT cells transfected with full-length, N-terminal or C-terminal α-COP shows that Nucleolin co-immunoprecipitates only full-length or C-terminal α-COP. (b) Schematic of eGFP-tagged Nucleolin and truncation mutants. Representative micrographs from transfected NSC-34 cells show that the N-terminal fragment of Nucleolin which contains the NLS is predominantly nuclear while the C-terminal fragment demonstrates more diffuse cytoplasmic localization. Cells were co-transfected with Lifeact Ruby to visualize cytoarchitecture and nuclei were visualized with DAPI (blue). (c) GFP-TRAP on whole cell lysate from HEK 293TT transfected with full-length, N-terminal or C-terminal eGFP-Nucleolin shows that α-COP clearly co-immunoprecipitates with full-length and C-terminal Nucleolin but not the N-terminal fragment.
The C-terminal dilysine of Nucleolin is required to bind α-COP
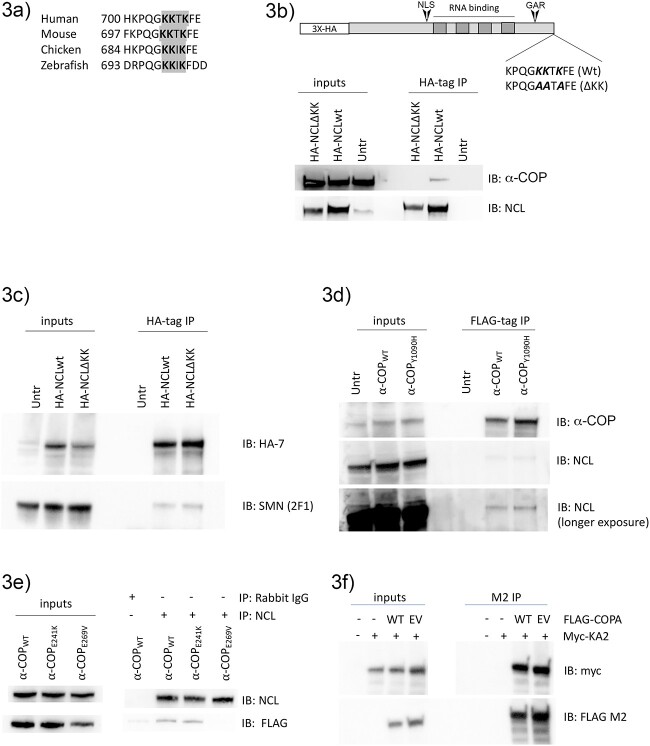
The C-terminus of Nucleolin among vertebrates contains a highly conserved dilysine motif (KxKxx) (Fig. 3a). C-terminal dilysine motifs, either KKxx or KxKxx are canonical COPI binding signals mediated by the N-terminal WD40 domains of β′-COP and α-COP respectively [33,45,46]. To determine whether the association between Nucleolin and α-COP is mediated by the C-terminal dilysine motif, we used site-directed mutagenesis to change the lysines to alanines (KKxKxx ➔ AAxAxx) using a previously published HA-tagged Nucleolin expression construct [47] hereafter referred to as NCLΔKK. Figure 3b shows a co-precipitation of HA-tagged NCLwt versus NCLΔKK in 293-TT cells. Both wildtype and ΔKK Nucleolin expressed and IP’d well, but only wildtype Nucleolin was able to co-IP endogenous α-COP. Since Nucleolin and α-COP are both able to co-IP SMN, we wondered if ΔKK could still co-IP SMN in the absence of α-COP. Figure 3c demonstrates that NCLΔKK is still able to co-IP SMN from 239TT whole cell lysates. We have previously reported a mutation in α-COP which eliminates SMN binding [17]. Similar to the finding that the interaction between Nucleolin and SMN does not require α-COP, experiments displayed in Fig. 3d demonstrate that when Flag-α-COP WT or Flag-α-COP Y1090H were IP’d from 293TT lysate, both were able to co-IP endogenous Nucleolin. Together, these findings indicate that the ability of α-COP to co-immunoprecipitate Nucleolin does not require SMN, and that the interaction between SMN and Nucleolin does not require α-COP. We propose that rather than a tripartite protein complex, Nucleolin is interacting with SMN in the nucleus and with α-COP in the cytoplasm.
Figure 3.
The C-terminal dilysine of Nucleolin mediates interaction with α-COP. (a) An alignment of the amino acid sequence of the Nucleolin C-terminus emphasizes a highly conserved dilysine motif (gray). (b) Schematic showing HA-tagged Nucleolin with either a wildtype or mutated C-terminal dilysine (HA-NCLWT vs HA-NCLΔKK). A representative Western blot from whole cell HEK 293TT lysate shows that in immunoprecipitations using HA antibody in cells transfected with HA-NCLWT or HA-NCLΔKK, endogenous α-COP only co-precipitates with HA-NCLWT. (c) In transfected HEK 293TT cells, both HA-NCLWT and HA-NCLΔKK can co-immunoprecipitate endogenous SMN. (d) A representative Western blot from HEK 293TT cells transfected with Flag-tagged α-COPWT or α-COPY1090H. Both wildtype and the Y1090H mutant co-immunoprecipitate endogenous SMN (bottom panel shows longer exposure). (e) Representative blot showing immunoprecipitation of endogenous Nucleolin from HEK 293TT cells expressing Myc/Flag-tagged α-COP variants. Both wild-type and E241K α-COP co-immunoprecipitate with endogenous Nucleolin, but E269V α-COP did not. (f) Representative blot showing immunoprecipitation of wildtype (WT) or E269V (EV) α-COP using an antibody against the FLAG tag from cells transfected with myc-KA2. Immunoblotting with FLAG and Myc-tag antibodies shows expression of both WT and EV α-COP as well as expression of Myc-KA2 in the inputs and shows that both WT and EV co-IP’d myc-KA2 while no myc-KA2 was detected in cells without MYC-FLAG-α-COP expression.
In humans, mutations in COPA result in an autosomal dominant condition referred to as COPA syndrome, characterized by arthritis and lung disease [48–50]. Subsequent work has revealed that expression of mutant α-COP causes aberrant trafficking and activation of STING [51–53]. In further dissecting the role of α-COP in STING processing, it was shown that the STING interacting protein Surf4 bound α-COP via a C-terminal dilysine motif. α-COP bearing the E241K mutation found in several COPA syndrome families displayed reduced binding to Surf4 compared with wildtype [54]. As the interaction between α-COP and Nucleolin appears mediated by the Nucleolin C-terminal dilysine, we wondered if α-COP-E241K would be able to co-IP Nucleolin, or whether its failure to bind dilysine motifs was specific to Surf4. This E241K mutation, however, is on the side of the dilysine-binding WD-40 domain instead of the middle where most of the primary interactions occur according to homology mapping and mutagenesis in yeast [55]). As a result, E241K likely only disrupts a subset of dilysine binding, not all. Yeast E273 (human E269) on the other hand, is in the central WD-40 barrel and directly contacts the dilysine motif. We therefore created an E269V human α-COP mutant to test whether Nucleolin binds the core WD-40 domain as is standard for general dilysine motifs. When endogenous Nucleolin was IP’d from cells expressing Myc/Flag-tagged α-COP wildtype, E241K, or E269V both wildtype and E241K α-COP co-IP’d, but the E269V mutant did not bind, indicating that this residue in the α-COP WD40 domain is critical for interaction with Nucleolin (Fig. 3e). To confirm that the E269V mutation did not interfere with the ability of α-COP to bind other domains, we transfected cells with Myc-tagged kainate receptor subunit 2 (KA2). This protein binds α-COP via an internal basic motif (RRRRR) and has been demonstrated to co-IP with α-COP [56]. Figure 3f shows that both wildtype (WT) and E269V (EV) α-COP were able to co-IP myc-KA2 indicating that this mutant is able to bind other COPI targets, but not the KxKxx domain of Nucleolin.
Binding to α-COP is not required for normal Nucleolin localization or mRNA binding
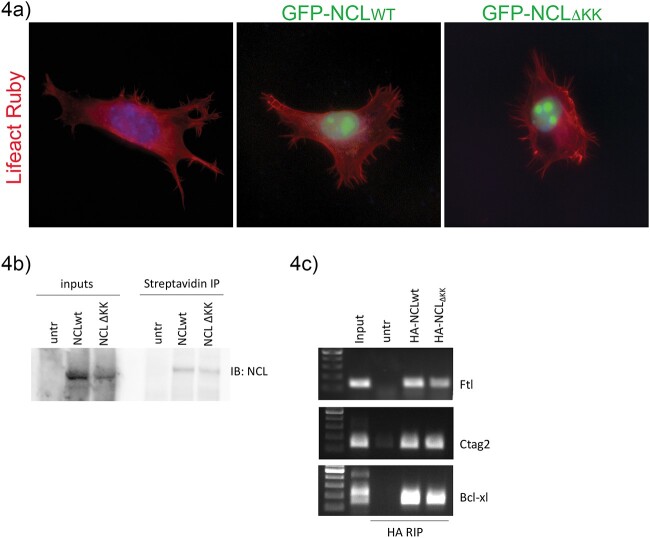
To determine if binding to α-COP was required for the normal nucleolar localization of Nucleolin, we subcloned the ΔKK into a previously published GFP-tagged Nucleolin expression vector [57]. Murine NSC-34 cells were plated on glass coverslips and transfected with GFP-NCLWT or GFP-NCLΔKK along with Lifeact ruby to visualize the cytoskeleton [58]. After 48 h, coverslips were fixed and immediately mounted in DAPI stained mounting media to visualize nuclei. Epifluorescent microscopy showed that both GFP-NCLWT and GFP-NCLΔKK displayed predominantly nucleolar localization and there were no significant differences in subcellular localization between the two (Fig. 4a). Cell surface localization of Nucleolin has been reported in numerous cell types but is notoriously difficult to visualize by fluorescent microscopy. To compare cell surface localization of WT and ΔKK Nucleolin, we transfected 239TT cells with HA-tagged Nucleolin and used cell impermeant biotin to label extracellular proteins. Figure 4b shows that comparable amounts of both HA-NCLWT and HA-NCLΔKK were detected at the cell surface when proteins were isolated using streptavidin beads. This is consistent with previous reports that delivery of Nucleolin to the cell surface was not affected by treatment with Brefeldin A, which inhibits COPI vesicle function by disrupting the Golgi apparatus [59]. Another assay of Nucleolin function related to its role as an RNA binding protein. When the mRNA associated with Nucleolin were characterized by RNA-IP in HeLa cells, Ferritin Light Chain (Ftl) and Cancer/Testis Antigen 2 (Ctag2) were among the most robust mRNA binding partners for Nucleolin [60]. Other groups have shown that Nucleolin binds Bcl-xl mRNA [61]. We used RT-PCR to confirm that Ftl, Ctag2 and Bcl-xl were expressed in 293TT cells (not shown), then used the HA-tagged Nucleolin expression vectors to examine mRNA binding by RNA-IP with HA-tag antibody. Endpoint PCR showed that both WT and ΔKK Nucleolin were able to co-IP Ftl, Ctag2 and Bcl-xl mRNA, indicating that α-COP binding is not required for Nucleolin to interact with these mRNA (Fig. 4c). Taken together, these data support the hypothesis that basal Nucleolin functions do not require interaction with α-COP at the C-terminal dilysine domain.
Figure 4.
Mutating the C-terminal dilysine motif does not impact normal Nucleolin function. (a) Representative micrographs of NSC-34 cells transfected with LifeAct Ruby (red) and eGFP-tagged Nucleolin. Nuclei were visualized with DAPI (blue). (b) Representative Western blot of HEK 293TT cells transfected with HA-NCLWT or HA-NCLΔKK shows equal cell-surface expression. (c) DNA gels showing RT-PCR from inputs or RIPs in untransfected control HEK 293TT compared to cells transfected with HA-NCLWT or HA-NCLΔKK.
Nucleolin bound mRNA show reduced association with a-COP after Nucleolin knockdown
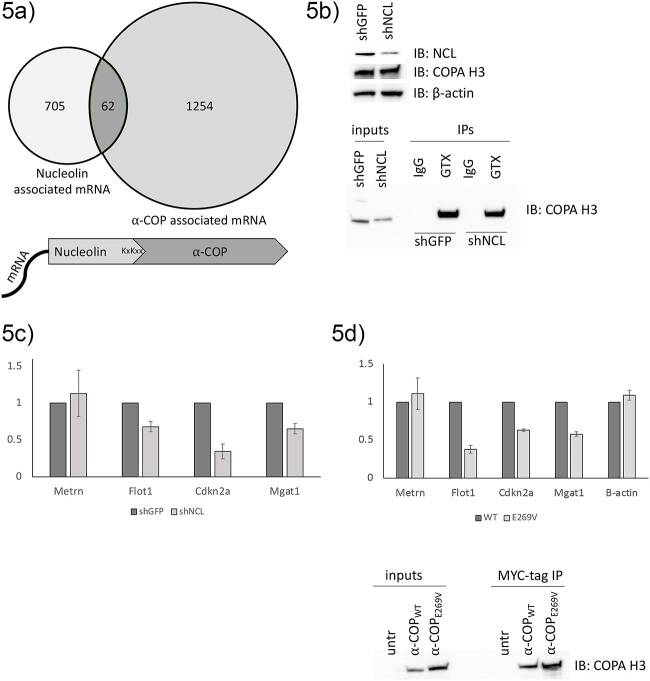
We propose that the interaction between Nucleolin and α-COP allows Nucleolin to act as a bridge between α-COP and its mRNA cargoes. Comparing our α-COP RIP-seq data set from motor neuron-like NSC-34 cells with the RIP-seq of Nucleolin in Hela cells to identify transcripts whose interaction with α-COP could be a result of Nucleolin binding, we find that roughly 9% of the reported Nucleolin associated transcripts were also detected in the α-COP RIP (Fig. 5a). In our original α-COP RIP-seq data set, we also detected significant overlap with mRNA reported to bind FMRP [26]. FMRP has also been reported to bind Nucleolin [62], so a Nucleolin/FMRP/α-COP complex could bridge an additional set of transcripts. To determine if the ability of α-COP to bind certain transcripts is Nucleolin dependent, we generated polyclonal cell lines stably expressing short hairpin RNA (shRNA) against either GFP (control) or the 3′-UTR of Nucleolin. Western blotting confirmed that Nucleolin was reduced at the protein level and α-COP protein levels remained unchanged, and we confirmed endogenous a-COP cleanly IP’d from both lines using a rabbit polyclonal antibody (Fig. 5b). Using this antibody against endogenous α-COP, we performed RNA-IP followed by quantitative RT-PCR for known Nucleolin interacting transcripts. Although the presence of Metrn mRNA in the α-COP RIP was unaffected by Nucleolin knockdown, three other primer sets demonstrated significant decreases in mRNA binding to α-COP after Nucleolin knockdown (Fig. 5c). To validate this result, we transfected cells with wildtype α-COP or the E269V mutant, which is unable to co-precipitate Nucleolin, our prediction being that since the E269V mutant is unable to bind Nucleolin, it would show reduced ability to immunoprecipitate Nucleolin-associated mRNA. We found that like RIPs performed in Nucleolin depleted cultures, quantitative RT-PCR demonstrated decreased binding of Flot1, Cdkn2, and Mgat1 in E269V RIPS compared to wildtype. Immunoprecipitation of β-actin mRNA, which has previously been shown to bind α-COP but not Nucleolin [15,35] showed no significant difference between α-COPWT and α-COPE269V RIPs.
Figure 5.
Nucleolin mediates binding of α-COP to some mRNA. (a) Venn diagram showing overlap of Nucleolin-associated mRNA in Hela cells with α-COP-associated mRNA in NSC-34 cells and proposed model of Nucleolin acting as an adapter between mRNA and α-COP. (b) Representative Western blot of whole cell lysates from stable shGFP and shNCL HEK 293TT lines shows decrease in Nucleolin at the protein level. Immunoprecipitation of endogenous α-COP with a rabbit polyclonal antibody (GTX) was approximately equal in both lines. (c) Quantitative RT-PCR of α-COP RIPs normalized to GAPDH. (d) Quantitative RT-PCR from RIPs comparing α-COPWT to α-COPE269V. A representative Western blot confirms successful expression and immunoprecipitation of the Myc-tagged α-COP.
Binding to Nucleolin is required for α-COP to fully support neurite outgrowth
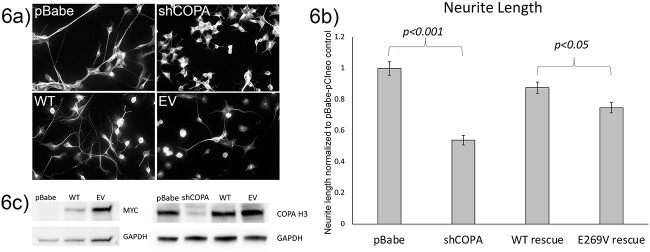
α-COP and Nucleolin have both been implicated in the regulation of neurite outgrowth. We have shown that knockdown of α-COP reduced the growth of both dendrites and axons in cortical neuron cultures and reduces neurite length in differentiated NSC-34 cells. [17] Similarly, treatment with Brefeldin A, which dissociates the COPI complex from the Golgi, inhibits axon outgrowth in primary hippocampal neuron cultures [63]. To determine whether binding Nucleolin is required for α-COP to support neurite outgrowth, we performed a complementation study in Copa knockdown motor neuron-like NSC-34 cells. As shown in representative micrographs of β-III tubulin-stained cultures, stable knockdown of murine Copa reduces neurite length. Expression of Myc-FLAG-α-COPWT in the Copa knockdown background significantly rescued neurite length compared to Copa knockdown alone, but Myc-FLAG-α-COPE269V was less able to rescue (post-hoc t-test p = 0.0101 compared to WT). Neurite lengths from three biological replicates are quantified in Fig. 6b. Stable expression of Myc-FLAG-α-COPWT and Myc-FLAG-α-COPE269V was confirmed by Western blotting with anti-Myc antibodies and reduction of α-COP reduction at the protein level in Copa knockdown cultures was confirmed by Western blotting with antibody against α-COP in whole cell lysates (Fig. 6c). These results demonstrate that the interaction between α-COP and Nucleolin has significance for the growth and maintenance of cytoarchitecture in a relevant cell type.
Figure 6.
Loss of Nucleolin binding reduces the ability of α-COP to promote neurite outgrowth. (a) Representative micrographs of β-III tubulin labeled NSC-34 cells from control (pBabe + pCI-neo) Copa knockdown (shCOPA) and Myc-FLAG-α-COP rescue lines (WT or EV) after 5 days of differentiation. (b) Normalized neurite length quantification from 3 biological replicates measured by a blinded observer shows significant decrease in neurite length following Copa knockdown (p < 0.001). Stable expression of α-COPWT significantly restored neurite length (p < 0001). Stable expression of α-COPE269V rescued neurite length to some degree, but neurites were significantly shorter compared to α-COPWT cultures (p = 0.01). Error bars represent standard error of the mean. (c) Representative Western blot of whole cell lysates confirming expression of Myc-FLAG-α-COPWT and Myc-FLAG-α-COPE269V. Representative Western blot of whole cell lysates with antibody against endogenous α-COP confirms reduced α-COP protein in shRNA lines and restoration to normal levels by stable expression of Myc-FLAG-α-COPWT and Myc-FLAG-α-COPE269V.
Discussion
We have shown here for the first time that the RNA binding protein Nucleolin can interact with the COPI coatomer member α-COP by means of a C-terminal dilysine motif, and that this interaction requires the glutamic acid at position 269 of the WD40 domain in α-COP (Fig. 3b and e). The interaction between Nucleolin and α-COP appears to be independent of Survival Motor Neuron (SMN) protein, which is known to bind to α-COP in the cytoplasm and Nucleolin in the nucleus (Fig. 3c and d) [15,27–29]. Immunoprecipitation of Nucleolin from fractionated cells and Promility Ligase Assay indicate that the bulk of its interaction with α-COP takes place in the cytoplasm (Fig. 1c and d). Given that Nucleolin is an RNA binding protein and α-COP has been shown to bind mRNA [26], we propose that Nucleolin acts as an adapter to allow association of α-COP with a subset of mRNA (Fig. 5). Nucleolin has also been shown to interact with the RNA binding protein FMRP [62]. When the mRNAs associated with α-COP were originally assessed by RNA-seq of formaldehyde cross-linked NSC-34 cells, it was noted that there was a highly significant overlap with a previously reported profile of transcripts associated with FMRP [26,64]. Although α-COP and FMRP have not yet been shown to interact directly, our results raise the possibility that Nucleolin could function as a scaffold by binding to both FMRP and α-COP, thus delivering both Nucleolin-associated transcripts as well as FMRP-associated mRNA to α-COP.
To further understand the potential biological ramifications of the Nucleolin/α-COP interaction, we must consider the biological pathways which are affected by both COPI coatomer and Nucleolin. We demonstrate in Fig. 6 that α-COP which is unable to bind Nucleolin is less able to support neurite outgrowth in the motor neuron-like NSC-34 cell line. Nucleolin appears to impact axon outgrowth by binding certain mRNAs and regulating their subcellular distribution. When Nucleolin was sequestered in the cell body of cultured dorsal root ganglion neurons (DRG), it resulted in increased axon outgrowth [35]. Interaction with members of the COPI coatomer is often used to restrict the subcellular localization of proteins, either for retention in the Endoplasmic Reticulum or to promote formation of heterodimers [56]. If the interaction with α-COP promotes the retention of Nucleolin in the cell body, this would promote axon outgrowth.
α-COP is part of a very stable heptameric COPI complex with a half-life of over 28 h [36]. When treated with chemicals or high salt, the COPI complex dissociates into a B-subcomplex comprised of α-COP, β′-COP, and ε-COP and an F-subcomplex containing β-COP, δ-COP/Arcn1, γ-COP, and ζ-COP [65–67]. Distinct cellular functions for the B-subcomplex have not been documented in intact cells, but there is evidence of COPI subunits functioning independently at the endosome where β-COP and ε-COP but not γ-COP were present [68]. Within the fully assembled COPI heptamer, the F-subcomplex interacts with activated Arf1 to bind the Golgi membrane [69], while the B-subcomplex confers cargo specificity [55]. We demonstrate here that endogenous Nucleolin appears to only co-precipitate the B-subcomplex as α-COP and ε-COP but not β-COP were present in co-immunoprecipitation experiments (Fig. 1a). This is the first report of an interaction being specific to the B-subcomplex rather than interacting with the complete COPI heptamer. This suggests that coatomer proteins are not strictly limited to functioning as COPI heptamers, but rather that the cytoplasmic B- and F-subcomplexes may have additional independent functions in the cell beyond traditional COPI-dependent trafficking.
Nucleolin is subject to multiple post-translational modifications that affect its function. Cytoplasmic Nucleolin is both N and O-glycosylated and fucosylated as it traverses the Golgi apparatus where COPI members are enriched [70,71]. While N-glycosylation is required for transport of Nucleolin to the plasma membrane and does not appear to be sensitive to COPI function [59], fucosylation of Nucleolin is required to modulate cell size and it is currently unknown whether this modification requires COPI [72]. Conversely, Nucleolin may be able to influence a-COP indirectly by mediating its splicing. A recent study of 5′ latent splice sites demonstrated that knockdown of Nucleolin resulted in retention of intron 15 in Copa transcripts [73].
Nucleolin and α-COP also share a small subset of known protein–protein interactions. As mentioned before, both Nucleolin and α-COP are able to bind SMN, but it was also shown by tandem affinity purification followed by mass spectrometry that both Nucleolin and α-COP interact with Spartin, the protein product of the SPG20 gene, mutations in which cause a form of hereditary spastic paraplegia [74]. This is particularly of interest as the paper used subcellular fractionation to demonstrate that endogenous Spartin was found in the mitochondrial fraction. While α-COP has been reported to deliver mRNA to the mitochondria under some conditions and to interact with Stasimon at the mitochondria-ER interface [24,75], Nucleolin has not been reported as localizing to the mitochondria.
The interaction between Nucleolin and α-COP may also be relevant in both development and aging. Nucleolin is required for normal craniofacial development in Zebrafish, and COPI coatomer mutations result in a distinct craniofacial defect in humans [76,77]. Nucleolin and α-COP have also been implicated in Alzheimer’s disease (AD) pathology, where mutations in α-COP and other COPI coatomer subunits are associated with increased risk of AD and Nucleolin is found to be decreased in late-stage AD brains and can also act as a microglial receptor for the amyloidogenic Aβ-42 and a modulator of amyloid precursor protein mRNA stability [78–82].
Material and methods
Cell culture and immunofluorescence
Hek 293TT and NSC-34 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% Fetal Bovine Serum (FBS) and Normocin (Invivogen catalog # ant-nr-2, 0.1 mg/ml). For transfections, plasmid DNA was combined with Polyethylenamine (PEI) in OPTI-MEM overnight [83]. For immunofluorescent microscopy experiments, NSC-34 cells were plated on poly-D-lysine coated coverslips and transfected with Lipofectamine 2000 at a 1:2 ratio. Coverslips were fixed in 4% paraformaldehyde 48 h post-transfection, then immediately mounted using Prolong Gold with DAPI. Images were captured on a Nikon Eclipse 80i at 40×.
Biotinylation, cell fractionation, and Nucleolin knockdown
For cell surface biotinylation experiments, cells were transfected with HA-tagged Nucleolin and labeled as described previously [78,84]. Briefly, 48 h post-transfection, cultures were treated with NHS-SS-Biotin for 1 h at 4°. The biotin was inactivated with Glycine, then cells were rinsed three times in cold PBS, lysed in RIPA buffer and cell surface proteins pulled down by tumbling on Streptavidin magnetic beads for 1 h. Nuclear/cytoplasmic fractionation was performed as described in [29] with the minor change of adding 0.075% NP40 detergent to buffer A. For generation of stable knockdown of Nucleolin or control cultures, HEK 293TT cells were transfected with shRNA (control SHC002 and Nucleolin 3′-UTR TRCN0000062283 both from Sigma). 48 h post-transfection cells were selected for expression of the shRNA by treatment with Puromycin (1 mg/ml) for 48 h. Nucleolin knockdown was verified by Western blot.
Antibodies
Nucleolin rabbit polyclonal (D4C70, Cell Signaling #14574). 1:3000 for Western blot, 1:500 for immunofluorescence, 1:300 for immunoprecipitation. Nucleolin mouse monoclonal (H-6, Santa Cruz #sc-55486). 1:1000 for Western blot, 1:100 for immunoprecipitation. Alpha-COP mouse monoclonal (H-3, Santa Cruz #sc-398099). 1:1000 for Western blotting, 1:100 for immunofluorescence. Alpha-Cop rabbit (GeneTex #GTX45827) 1:1000 for RIP. Survival Motor Neuron protein mouse monoclonal (2F1, Cell Signaling #12976). 1:3000 for Western blotting. e-COP mouse monoclonal (Santa Cruz #sc-133194). 1:1000 for Western blotting. b-COP (Abcam #ab2899). 1:5000 for Western blotting.
Immunoprecipitations
For Immunoprecipitation, cells were lysed in polysome lysis buffer (PLB-10 mM Hepes pH 7.5, 5 mM MgCl2, 100 mM KCl, 0.5% NP40, 1 mM DTT, Protease inhibitor cocktail (Sigma S8830). Soluble lysate was incubated with BSA pre-blocked magnetic protein A/G beads for 3 h at 4C. For immunoprecipitations in the presence of RNase, 1 ml cell lysates were incubated with 5 ul 20 mg/ml RNase A at 30C for 15 min before addition of beads and antibody. RNA-immunoprecipitations (RIPs) were performed as described in [85] with slight modifications. Magnetic protein A/G beads were blocked with 2% bovine serum albumin and 0.8 mM random oligo for 1 h before use. RIPs were carried out with end-over-end rotation at 4° for 3 h. After washing, the beads were resuspended in 100 ml PLB and treated with Proteinase K for 45 min to release protein/RNA complexes. Total RNA was then extracted using the RNeasy kit (Qiagen #74004) with on-column DNAse treatment (Qiagen #79254). Total RNA was converted to cDNA using the iScript cDNA synthesis kit (BioRad #1708890). For end-point PCR, 1 ml of cDNA was amplified using Bullseye Premium R-taq 2× mix (Midwest Scientific #BE180303). For quantitative PCR, 1 ml of cDNA was amplified using iQ SYBR green supermix (BioRad #1708880). RT-PCR primers for Ftl, Ctag2, BCL-XL, Metrn, Flot1, Cdkn2a, Mgat1, and b-actin were used as described [29,60,61].
Plasmid construction
The C-terminal dilysine motif of HA-Nucleolin was mutated to AxAxx by site-directed mutagenesis in two steps, using the following primers: KxA Forward 5′ GAAAGAAGACGGCATTCGAATAGG AATTC 3′ KxA Reverse 5′ GAATCCCTATTCGAATGCCGTCTTCTTTC 3′ AxA Forward 5′ CACAAGCCACAAGGAGCAAAGACTGCATTCGAATAG 3′ AxA Reverse 5′ CTATTCGA ATGCAGTCTTTGCTCCTTGT GGCTTGTG 3′. To generate full-length GFP-tagged Nucleolin wildtype or AxAxx, the HA-tagged Nucleolin was subcloned by PCR using the following primers, then ligated into the SalI and EcoRI sites of peGFP-C1. Forward NCL GFP 5′ GATCGAATTCTATGGTGAAGCTCGATC 3′ NCL reverse wt-SalI 5′ GTACGTCG ACCTATTCAAACTTCG 3′ NCL reverse AAxA-SalI 5′ GTACGTCGACCTATTCGAATGCCGTC 3′. To generate the N-terminal fragment of GFP-Nucleolin (Addgene plasmid #28176), the following primers were used for to produce a PCR product, which replaced full-length Nucleolin by ligation into the KpnI/BamHI sites of the vector. N-terminal truncation Forward 5′ CAAGCTTGGTACCATGGTGAAGCTCGCGAA 3′ N-terminal truncation Reverse 5′ GAACGTTGGATCCATATCAGACAGG CTCTTC 3′. For the C-terminal truncation, the following primers were used to PCR amplify the appropriate fragment which again was cloned into KpnI/BamHI to replace the full-length Nucleolin in the vector with the C-terminal half of the protein beginning at RRM1. C-terminal truncation Forward 5′ GCACGTGGTACCTTCAATCTCTTTGTTG 3′ C-terminal truncation Reverse 5′ CTAGATCCGGTGGATCCAGACATGATAAG 3′. pCDNA6-human a-COP-myc-FLAG (Origene #RC412199) was used as a template for site-directed mutagenesis and generation of truncation mutants. Template was amplified with Pfu polymerase 18×, digested with Dpn1 restriction enzyme, and transformed into DH5a. Colonies were screened by restriction enzyme digestion. Primers for E269V mutation also introduced a silent mutation Sal1 site to allow for easy colony screening: Forward: CAGCAATTCTGTCGACAAGAGTATTC and Reverse: GAATACTCTTGTCGACAGAATTGCTG. α-COP 813-1224 C-terminal truncation was PCR amplified with hacop-Mlu-R (GATT ACGCGT GCGAAACTGCAG) and AsiS1-haCOP-813-Fb (atta GCGATCGCC ATG gta tcc aaa gg). Vector and PCR products were digested with Mlu1 and AsiSI, gel purified, ligated, and transformed into DH5a. α-COP 1–812 was generated through insertion of ligated Mfe-acop812-Not-F/R (AATTGGCCTTTATTGACT ACGCGT ACGC, GGCCGCGT ACGCGT AGTCAATAAAGGCC) primers into Mfe/Not1 digested pCDNA6-human α-COP -myc-FLAG vector. E241K α-COP was a gift from Dr Anthony Shum.
Proximity ligation assay
Murine neuroblastoma or NSC-34 cells were plated on PDL-coated coverslips and fixed in 4% PFA followed by permeabilization in 0.2% Triton-X for 30 min. PLA was performed as directed using the DuoLink kit (DUO92101). Antibodies and dilutions were as follows: α-COP (SC H-3) 1:100, NCL (D4C70) 1:500, β′-COP (Bethyl labs A304-522A) 1:100.
NSC-34 neurite outgrowth assay
Motor neuron-like NSC-34 cells were transfected with pBabe-puro or a combination of 3 shRNA targeting the murine Copa 3′-UTR (TRCN0000113147, TRCN0000313321, TRCN0000349438) and selected with puromycin. Once Copa knockdown was established, control and knockdown cultures were transfected with either pCI-neo or Myc-FLAG-α-COP followed by selection with G418. Western blot with anti-myc antibody confirmed the stable expression of α-COPWT and α-COPE269V while blotting with antibody for α-COP confirmed that shRNA reduced α-COP at the protein level and expression of returned the α-COP to levels comparable to the pBabe-pCI-neo control line. For differentiation, cells were plated on poly-D-lysine (150 μg/ml) coated coverslips in DMEM/F12 with 1% FBS and retinoic acid (10 μM) and grown for 5 days before fixation in 4% PFA. Neurites were visualized by immunofluorescence with antibody against β-III tubulin. Images were captured and neurite length measured using the ImageJ (simple neurite tracer) by a blinded observer.
Funding
This work was funded by support from the National Institute of Neurological Disorders and Stroke (1RO1NS082284-01A1). Ms Iurillo was supported by the Indiana University Medical Student Program for Research and Scholarship (IMPRS) funded by UL1TR002529 from the National institutes of Health.
Conflict of interest statement: None declared.
Data availability
All plasmids and cell lines described here will be made available upon request.
Contributor Information
Sara K Custer, Dermatology, Indiana University School of Medicine, 545 Barnhill Drive, Emerson Hall 139, Indianapolis, IN 46202, United States.
Timra Gilson, Dermatology, Indiana University School of Medicine, 545 Barnhill Drive, Emerson Hall 139, Indianapolis, IN 46202, United States.
Jacob W Astroski, Dermatology, Indiana University School of Medicine, 545 Barnhill Drive, Emerson Hall 139, Indianapolis, IN 46202, United States.
Siddarth R Nanguneri, Dermatology, Indiana University School of Medicine, 545 Barnhill Drive, Emerson Hall 139, Indianapolis, IN 46202, United States.
Alyssa M Iurillo, Indiana University School of Medicine, 340 West 10th St, Indianapolis, IN 46202, United States.
Elliot J Androphy, Dermatology, Indiana University School of Medicine, 545 Barnhill Drive, Emerson Hall 139, Indianapolis, IN 46202, United States.
References
- 1. Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 2005;48:885–95. [DOI] [PubMed] [Google Scholar]
- 2. Tisdale S, Lotti F, Saieva L et al. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3′-end formation of histone mRNAs. Cell Rep 2013;5:1187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Donlin-Asp PG, Fallini C, Campos J et al. The survival of motor neuron protein acts as a molecular chaperone for mRNP assembly. Cell Rep 2017;18:1660–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jangi M, Fleet C, Cullen P et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc Natl Acad Sci U S A 2017;114:E2347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Custer SK, Gilson TD, Li H et al. Altered mRNA splicing in SMN-depleted motor neuron-like cells. PLoS One 2016;11:e0163954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huo Q, Kayikci M, Odermatt P et al. Splicing changes in SMA mouse motoneurons and SMN-depleted neuroblastoma cells: evidence for involvement of splicing regulatory proteins. RNA Biol 2014;11:1430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez G, Dury AY, Murray LM et al. A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet 2013;22:668–84. [DOI] [PubMed] [Google Scholar]
- 8. Lauria F, Bernabo P, Tebaldi T et al. SMN-primed ribosomes modulate the translation of transcripts related to spinal muscular atrophy. Nat Cell Biol 2020;22:1239–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rathod R, Havlicek S, Frank N et al. Laminin induced local axonal translation of β-actin mRNA is impaired in SMN-deficient motoneurons. Histochem Cell Biol 2012;138:737–48. [DOI] [PubMed] [Google Scholar]
- 10. Fallini C, Donlin-Asp PG, Rouanet JP et al. Deficiency of the survival of motor neuron protein impairs mRNA localization and local translation in the growth cone of motor neurons. J Neurosci 2016;36:3811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rihan K, Antoine E, Maurin T et al. A new cis-acting motif is required for the axonal SMN-dependent Anxa2 mRNA localization. RNA 2017;23:899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kye MJ, Niederst ED, Wertz MH et al. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet 2014;23:6318–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng C, Reinhard S, Hennlein L et al. Impaired dynamic interaction of axonal endoplasmic reticulum and ribosomes contributes to defective stimulus-response in spinal muscular atrophy. Transl Neurodegener 2022;11:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beck R, Rawet M, Wieland FT et al. The COPI system: molecular mechanisms and function. FEBS Lett 2009;583:2701–9. [DOI] [PubMed] [Google Scholar]
- 15. Peter CJ, Evans M, Thayanithy V et al. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet 2011;20:1701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Custer SK, Todd AG, Singh NN et al. Dilysine motifs in exon 2b of SMN protein mediate binding to the COPI vesicle protein alpha-COP and neurite outgrowth in a cell culture model of spinal muscular atrophy. Hum Mol Genet 2013;22:4043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Custer SK, Gilson T et al. Alpha-COP binding to the survival motor neuron protein SMN is required for neuronal process outgrowth. Hum Mol Genet 2015;24:7295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Custer SK, Astroski JW, Li HX et al. Interaction between alpha-COP and SMN ameliorates disease phenotype in a mouse model of spinal muscular atrophy. Biochem Biophys Res Commun 2019;514:530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chuang CF, King CE, Ho BW et al. Unbiased proteomic study of the axons of cultured rat cortical neurons. J Proteome Res 2018;17:1953–66. [DOI] [PubMed] [Google Scholar]
- 20. Cagnetta R, Frese CK, Shigeoka T et al. Rapid cue-specific Remodeling of the nascent axonal proteome. Neuron 2018;99:29–46.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C, Zhang Y, Levin AM et al. Distal axonal proteins and their related MiRNAs in cultured cortical neurons. Mol Neurobiol 2019;56:2703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de La Vega LA, Stockert RJ. The cytoplasmic coatomer protein COPI. A potential translational regulator. J Biol Chem 1999;274:31135–8. [DOI] [PubMed] [Google Scholar]
- 23. Trautwein M, Dengjel J, Schirle M et al. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol Biol Cell 2004;15:5021–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zabezhinsky D, Slobodin B, Rapaport D et al. An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep 2016;15:540–9. [DOI] [PubMed] [Google Scholar]
- 25. Bi J, Tsai NP, Lu HY et al. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc Natl Acad Sci U S A 2007;104:13810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Todd AG, Lin H, Ebert AD et al. COPI transport complexes bind to specific RNAs in neuronal cells. Hum Mol Genet 2013;22:729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lefebvre S, Burlet P, Viollet L et al. A novel association of the SMN protein with two major non-ribosomal nucleolar proteins and its implication in spinal muscular atrophy. Hum Mol Genet 2002;11:1017–27. [DOI] [PubMed] [Google Scholar]
- 28. Shafey D, Boyer JG, Bhanot K et al. Identification of novel interacting protein partners of SMN using tandem affinity purification. J Proteome Res 2010;9:1659–69. [DOI] [PubMed] [Google Scholar]
- 29. Gabanella F, Barbato C, Fiore M et al. Fine-tuning of mTOR mRNA and Nucleolin complexes by SMN. Cell 2021;10. 10.3390/cells10113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Masuzawa T, Oyoshi T. Roles of the RGG domain and RNA recognition motif of Nucleolin in G-Quadruplex stabilization. ACS Omega 2020;5:5202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhai LY, Liu JF, Zhao JJ et al. Targeting the RNA G-Quadruplex and protein Interactome for antiviral therapy. J Med Chem 2022;65:10161–82. [DOI] [PubMed] [Google Scholar]
- 32. Patel P, Buchanan CN, Zdradzinski MD et al. Intra-axonal translation of Khsrp mRNA slows axon regeneration by destabilizing localized mRNAs. Nucleic Acids Res 2022;50:5772–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doron-Mandel E, Koppel I, Abraham O et al. The glycine arginine-rich domain of the RNA-binding protein nucleolin regulates its subcellular localization. EMBO J 2021;40:e107158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terenzio M, Koley S, Samra N et al. Locally translated mTOR controls axonal local translation in nerve injury. Science 2018;359:1416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perry RB, Rishal I, Doron-Mandel E et al. Nucleolin-mediated RNA localization regulates neuron growth and cycling cell size. Cell Rep 2016;16:1664–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lowe M, Kreis TE. In vivo assembly of coatomer, the COP-I coat precursor. J Biol Chem 1996;271:30725–30. [DOI] [PubMed] [Google Scholar]
- 37. Borer RA, Lehner CF, Eppenberger HM et al. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989;56:379–90. [DOI] [PubMed] [Google Scholar]
- 38. Chen CY, Gherzi R, Andersen JS et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev 2000;14:1236–48. [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang Y, Xu XS, Russell JE. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Mol Cell Biol 2006;26:2419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eugster A, Frigerio G, Dale M et al. The alpha- and beta'-COP WD40 domains mediate cargo-selective interactions with distinct di-lysine motifs. Mol Biol Cell 2004;15:1011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsia KC, Hoelz A. Crystal structure of alpha-COP in complex with epsilon-COP provides insight into the architecture of the COPI vesicular coat. Proc Natl Acad Sci U S A 2010;107:11271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Creancier L, Prats H, Zanibellato C et al. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell 1993;4:1239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edwards TK, Saleem A, Shaman JA et al. Role for nucleolin/Nsr1 in the cellular localization of topoisomerase I. J Biol Chem 2000;275:36181–8. [DOI] [PubMed] [Google Scholar]
- 44. Schmidt-Zachmann MS, Nigg EA. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci 1993;105:799–806. [DOI] [PubMed] [Google Scholar]
- 45. Spang A. Traffic COPs: rules of detection. EMBO J 2013;32:915–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma W, Goldberg J. Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J 2013;32:926–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hanakahi LA, Dempsey LA, Li MJ et al. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci U S A 1997;94:3605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kumrah R, Mathew B, Vignesh P et al. Genetics of COPA syndrome. Appl Clin Genet 2019;12:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watkin LB, Jessen B, Wiszniewski W et al. COPA mutations impair ER-Golgi transport and cause hereditary autoimmune-mediated lung disease and arthritis. Nat Genet 2015;47:654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deng Z, Law CS, Ho FO et al. A defect in Thymic tolerance causes T cell-mediated autoimmunity in a murine model of COPA syndrome. J Immunol 2020;204:2360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kato T, Yamamoto M, Honda Y et al. Augmentation of stimulator of interferon genes-induced type I interferon production in COPA syndrome. Arthritis Rheumatol 2021;73:2105–15. [DOI] [PubMed] [Google Scholar]
- 52. Lepelley A, Martin-Niclos MJ, Le Bihan M et al. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J Exp Med 2020;217:e20200600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Steiner A, Hrovat-Schaale K, Prigione I et al. Deficiency in coatomer complex I causes aberrant activation of STING signalling. Nat Commun 2022;13:2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng Z, Chong Z, Law CS et al. A defect in COPI-mediated transport of STING causes immune dysregulation in COPA syndrome. J Exp Med 2020;217. 10.1084/jem.20201045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jackson LP, Lewis M, Kent HM et al. Molecular basis for recognition of dilysine trafficking motifs by COPI. Dev Cell 2012;23:1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vivithanaporn P, Yan S, Swanson GT. Intracellular trafficking of KA2 kainate receptors mediated by interactions with coatomer protein complex I (COPI) and 14-3-3 chaperone systems. J Biol Chem 2006;281:15475–84. [DOI] [PubMed] [Google Scholar]
- 57. Takagi M, Absalon MJ, McLure KG et al. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005;123:49–63. [DOI] [PubMed] [Google Scholar]
- 58. Riedl J, Crevenna AH, Kessenbrock K et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods 2008;5:605–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hovanessian AG, Puvion-Dutilleul F, Nisole S et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res 2000;261:312–28. [DOI] [PubMed] [Google Scholar]
- 60. Abdelmohsen K, Tominaga K, Lee EK et al. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res 2011;39:8513–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang J, Tsaprailis G, Bowden GT. Nucleolin stabilizes Bcl-X L messenger RNA in response to UVA irradiation. Cancer Res 2008;68:1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taha MS, Nouri K, Milroy LG et al. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS One 2014;9:e91465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jareb M, Banker G. Inhibition of axonal growth by brefeldin a in hippocampal neurons in culture. J Neurosci 1997;17:8955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol 1999;19:7925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fiedler K, Veit M, Stamnes MA et al. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science 1996;273:1396–9. [DOI] [PubMed] [Google Scholar]
- 66. Lowe M, Kreis TE. In vitro assembly and disassembly of coatomer. J Biol Chem 1995;270:31364–71. [DOI] [PubMed] [Google Scholar]
- 67. Pavel J, Harter C, Wieland FT. Reversible dissociation of coatomer: functional characterization of a beta/delta-coat protein subcomplex. Proc Natl Acad Sci U S A 1998;95:2140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aniento F, Gu F, Parton RG et al. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol 1996;133:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu X, Breitman M, Goldberg J. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell 2012;148:530–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Carpentier M, Morelle W, Coddeville B et al. Nucleolin undergoes partial N- and O-glycosylations in the extranuclear cell compartment. Biochemistry 2005;44:5804–15. [DOI] [PubMed] [Google Scholar]
- 71. Aldi S, Della Giovampaola C, Focarelli R et al. A fucose-containing O-glycoepitope on bovine and human nucleolin. Glycobiology 2009;19:337–43. [DOI] [PubMed] [Google Scholar]
- 72. Palumberi D, Aldi S, Ermini L et al. RNA-mediated gene silencing of FUT1 and FUT2 influences expression and activities of bovine and human fucosylated nucleolin and inhibits cell adhesion and proliferation. J Cell Biochem 2010;111:229–38. [DOI] [PubMed] [Google Scholar]
- 73. Shefer K, Boulos A, Gotea V et al. A novel role for nucleolin in splice site selection. RNA Biol 2022;19:333–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Milewska M, McRedmond J, Byrne PC. Identification of novel spartin-interactors shows spartin is a multifunctional protein. J Neurochem 2009;111:1022–30. [DOI] [PubMed] [Google Scholar]
- 75. Van Alstyne M, Lotti F, Dal Mas A et al. Stasimon/Tmem41b localizes to mitochondria-associated ER membranes and is essential for mouse embryonic development. Biochem Biophys Res Commun 2018;506:463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dash S, Trainor PA. Nucleolin loss of function leads to aberrant fibroblast growth factor signaling and craniofacial anomalies. Development 2022;149. 10.1242/dev.200349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Izumi K, Brett M, Nishi E et al. ARCN1 mutations cause a recognizable craniofacial syndrome due to COPI-mediated transport defects. Am J Hum Genet 2016;99:451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Astroski JW, Akporyoe LK, Androphy EJ et al. Mutations in the COPI coatomer subunit alpha-COP induce release of Abeta-42 and amyloid precursor protein intracellular domain and increase tau oligomerization and release. Neurobiol Aging 2021;101:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bettayeb K, Hooli BV, Parrado AR et al. Relevance of the COPI complex for Alzheimer's disease progression in vivo. Proc Natl Acad Sci U S A 2016;113:5418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ozawa D, Nakamura T, Koike M et al. Shuttling protein nucleolin is a microglia receptor for amyloid beta peptide 1-42. Biol Pharm Bull 2013;36:1587–93. [DOI] [PubMed] [Google Scholar]
- 81. Garcia-Esparcia P, Sideris-Lampretsas G, Hernandez-Ortega K et al. Altered mechanisms of protein synthesis in frontal cortex in Alzheimer disease and a mouse model. Am J Neurodegener Dis 2017;6:15–25. [PMC free article] [PubMed] [Google Scholar]
- 82. Rajagopalan LE, Westmark CJ, Jarzembowski JA et al. hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res 1998;26:3418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Longo PA, Kavran JM, Kim MS et al. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol 2013;529:227–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Custer SK, Foster JN, Astroski JW et al. Abnormal Golgi morphology and decreased COPI function in cells with low levels of SMN. Brain Res 2019;1706:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jayaseelan S, Doyle F, Currenti S et al. RIP: an mRNA localization technique. Methods Mol Biol 2011;714:407–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All plasmids and cell lines described here will be made available upon request.