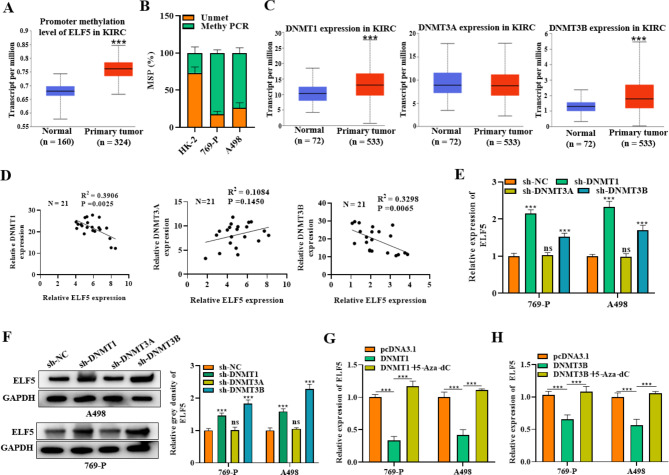
Fig. 5.
DNMTs promote hypermethylation of ELF5 in RCC. (A) UALCAN database was utilized to predict ELF5 methylation status in KIRC samples and normal samples. (B) MSP assay was utilized to detect ELF5 methylation in HK-2, 769-P, and A498. (C) UALCAN database was utilized for predicting DNMT1, DNMT3A, and DNMT3B expression in KIRC. (D) The correlation between ELF5 and DNMT1/DNMT3A/DNMT3B in RCC patient tissues (n = 21). (E-F) ELF5 expression was determined by RT-qPCR and western blot in cells when DNMT1, DNMT3A, or DNMT3B was silenced. (G-H) ELF5 level was determined through RT-qPCR when DNMT1/DNMT3B was overexpressed, and 5-Aza‐dC was added in cells. Two-tailed Student’s t test was performed. The error bars represent SD. ns means no significance, ***p < 0.001