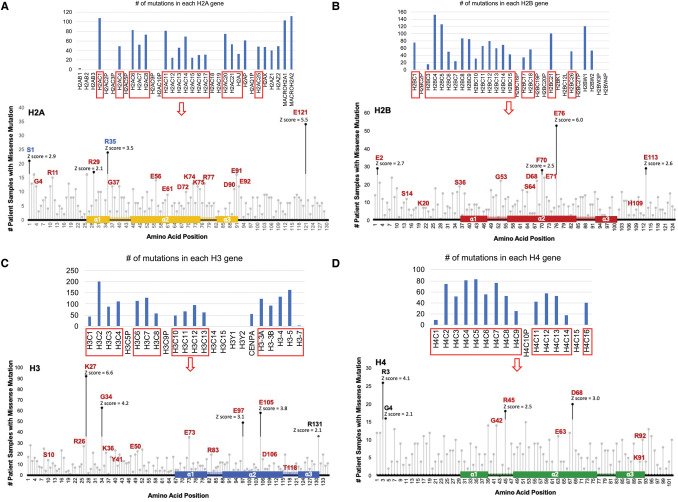
Figure 2. Survey of the most frequent core histone mutations in cancer patient samples.
A cross cancer mutation summary was performed using the cBioPortal to search a total of 29 836 non-redundant patient samples across all cancer types, excluding sample not profiled for all histone genes. The number of patients reported to have a missense mutation in the nucleosome core histones H2A, H2B, H3 and H4 at each amino acid residue position across well conserved histone paralogs was graphed by lollipop plot to depict the location of the cumulative missense mutations found in: H2A (A), H2B (B), H3 (C), and H4 (D). Alignment of histone proteins was performed in SnapGene v7.0.1. A Z-score was calculated by subtracting the number of mutations at a particular amino acid from the mean number of mutations across all amino acids and dividing the result by the standard deviation across all amino acids to identify the amino acids most frequently found mutated in each histone. Amino acid positions indicated by black dots are at least two standard deviations above the average number of mutations across all residues. Amino acids displayed in red were observed to have functional defects when mutated by Bagert et al. [27]. Amino Acids in blue were reported to have a similar polymorphism frequency between the general population (dbSNP) and cancer patient (cBioPortal) databases [25].