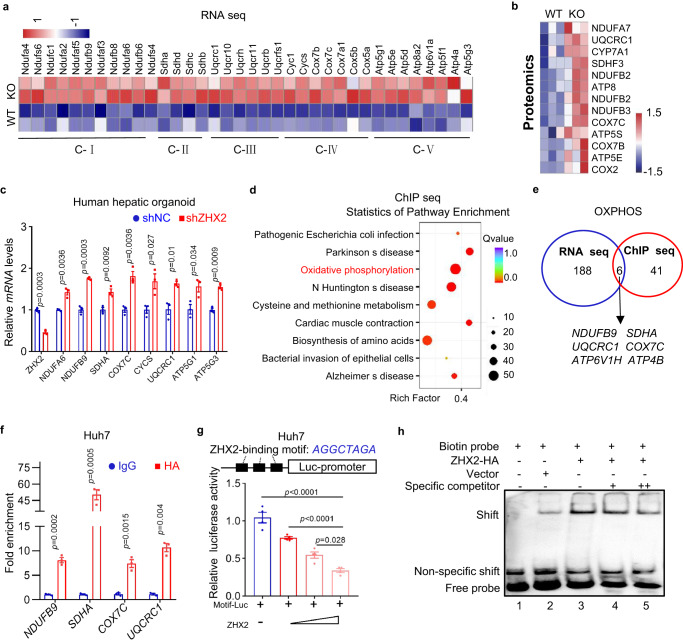
Fig. 5. ZHX2 transcriptionally represses ETC gene expression by binding to promoter regions.
a, b Heatmap from RNA-seq data and proteomics data of Zhx2-KOhep and Zhx2-WT mice livers at 48 h after 2/3 PHx. a Heatmap based on mRNA levels of mitochondrial complexes genes. b Heatmap showed proteins levels related to energy metabolism. Red and blue represent the increase and decrease of gene expression levels, respectively. c Relative mRNA levels of ETC genes were measured in human hepatic organoid cells transfected with LV-shZHX2 by RT-qPCR. Data are presented as mean ± sd (two-tailed Student’s t-test. n = 3 biologically independent samples). d ChIP-seq was performed with HA antibody using cell lysate from ZHX2-HA overexpressing Huh7 cells. KEGG analysis showed top enriched gene sets based on ChIP-seq data. e Venn diagram showed genes in OXPHOS pathway obtained from RNA-seq and ChIP-seq. f ChIP assay was performed with anti-HA antibody, IgG as control, using Huh7 cells transfected with ZHX2-HA. ZHX2 occupied at indicated genes were quantified by qPCR. Data are presented as mean ± sd (two-tailed Student’s t-test. n = 3 biologically independent samples). g The luciferase reporter vector containing 3 putative ZHX2-binding motifs was co-transfected with a gradient dose of ZHX2-HA vectors in Huh7 cells. The reporter promoter activities were displayed. Data are presented as mean ± sd. (One-way ANOVA with Tukey’s test. n = 4 biologically replicates). h EMSA were performed with nuclear extracts from Huh7 cells transfected with ZHX2-HA and control vectors. Biotin-labeled ZHX2-binding motif as probe and non-labeled motif as specific competitors.