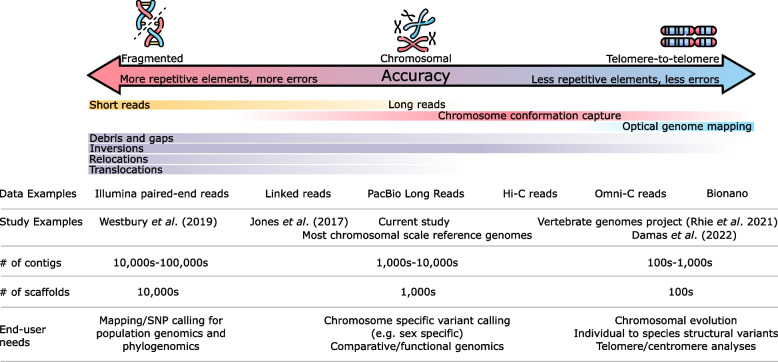
Fig. 4.
Hypothesized conceptual summary of reference genome accuracy, with specific reference to the construction of monodontid reference genomes. Note, in general, the percentage of repeat elements for a given genome skew accuracy in an inverse fashion. Errors are expected to diminish as more datatypes are combined, particularly those that resolve chromosomal scale ordering. We hypothesize debris is replaced, to a degree, by inversions, the latter of which feature a “long-tail” of persistence as datatypes are improved. End-user needs listed here may be carried forward with improved accuracy, but not backwards. Note, assembly accuracy approaches telomere-to-telomere assemblies, but fully resolving these regions remains the exception rather than the rule [6]. We also assume accuracy scales with the genetic distances proxied by the data types listed here, and acknowledge that assembly and scaffolding methods represent another variable in assembly accuracy not captured here. Images: Flaticon.com