Fig. 3 |. Biochemistry of protease-based protein and peptide ligations.
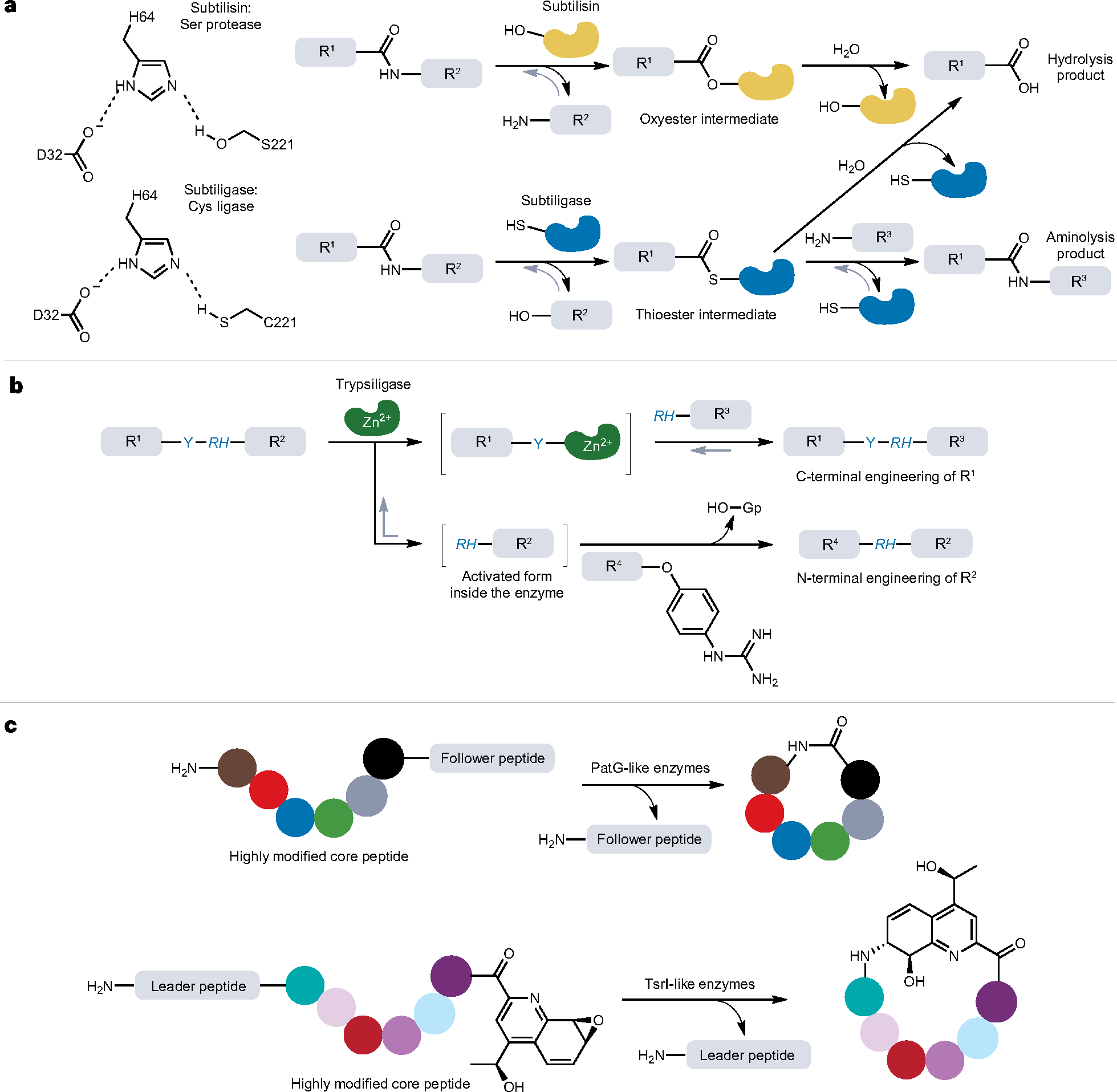
a, Biochemical mechanisms of subtilisin and subtiligase-mediated protein and peptide ligation. The catalytic triad of subtilisin is Asp–His–Ser (D32–H64–S221) and that of subtiligases is Asp–His–Cys (D32–H64–C221). For subtiligase, aminolysis is favoured over hydrolysis, thereby enabling the enzyme to support protein ligation. b, Trypsiligase-mediated protein ligation and its applications. The Y–RH sequence can be recognized and cleaved by trypsiligase. Guanidinophenyl (Gp) is a good leaving group that activates the C terminus of acyl donor, R4. R1–R4 denotes peptide or protein sequences throughout (a,b). c, Macrolactonization catalysed by protease-derived macrocyclases, PatG and TsrI, involved in microbial biosynthetic pathways. These enzymes catalyse not only the removal of signal peptides (such as leader or follower peptides) but also the intramolecular peptide bond formation between the N and C termini, as in the case of peptide macrolactonization. The coloured circles represent individual amino acid residues after post-translational modifications.
