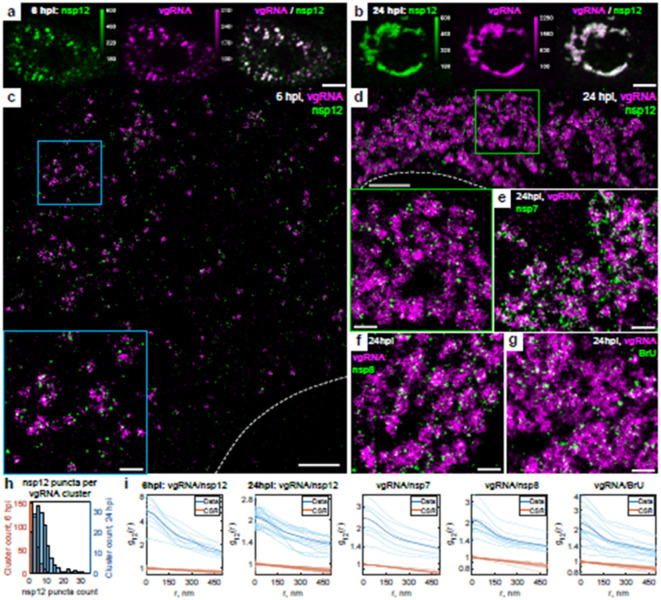
Fig. 3: Association of SARS-CoV-2 replication enzyme with vgRNA clusters.
a-b, Representative confocal images of SARS-CoV-2 infected cells display DL colocalization between nsp12, the catalytic subunit of RdRp (green) and vgRNA (magenta) at both 6 hpi (a) and 24 hpi (b). c-d, Representative SR images of SARS-CoV-2 infected cells indicate nanoscale association between nsp12 and vgRNA at both 6 hpi (c) and 24 hpi (d). Insets show magnified images of corresponding regions in the colored boxes. e-f, Representative SR images of vgRNA with nsp7 (e) or nsp8 (f) in the perinuclear regions of SARS-CoV-2 infected cells indicate association of nsp7 and nsp8 with vgRNA clusters. g, Representative SR image of vgRNA with newly synthesized viral RNAs labeled by BrU in a SARS-CoV-2 infected cell indicates localization of newly synthesized viral RNAs within the perinuclear clusters of vgRNA. h, Number of nanoscale puncta of nsp12 per vgRNA cluster. i, Bivariate pair-correlation functions for vgRNA and nsp12, nsp7, nsp8 and newly transcribed viral RNA labeled with BrU peak at r = 0 nm indicating association between these target pairs. Scale bars, 10 μm (a-b), 2 μm (c-d), 500 nm (e-g and insets in c-d). Dashed lines in c and d indicate the edge of the cell nucleus.