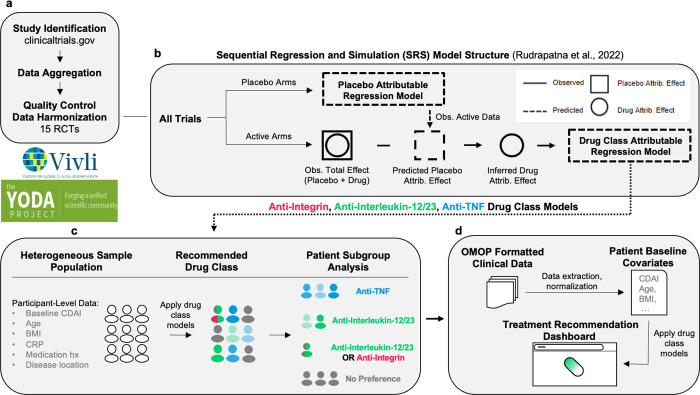
Figure 1: Overview.
A. Clinical trials were found using clinicaltrials.gov and sought for retrieval on the YODA and Vivli platforms. Individual participant data (IPD) from trials that collected CDAI scores at week 6 visits were then aggregated and harmonised. B. Using sequential regression and simulation, a method for normalising clinical trial data against a common placebo rate, a placebo-attributable model and three drug-attributable models - anti-integrin, anti-interleukin-12/23 and anti-TNF - were developed. Disease activity reduction was partitioned into placebo attributable (square) and drug-attributable (circle) effects based on baseline covariates (age, sex, BMI, etc.). IPD (solid lines) were used to predict or simulate data (dashed lines). C. The drug-attributable models were utilised to simulate patient-level outcomes post-treatment (counterfactuals). Pairwise t-tests (p < 0.05) were conducted to compare and rank the mean responses for all drug classes - anti-integrin vs anti-interleukin-12/23, anti-integrin vs anti-TNF, and anti-interleukin-12/23 vs anti-TNF - and assign patients into one of seven subgroup memberships (see Table 3). D. Lastly, the models were re-packaged into a prototype decision support tool that uses manual inputs and optionally, OMOP-formatted data, to recommend treatments for individual patients.