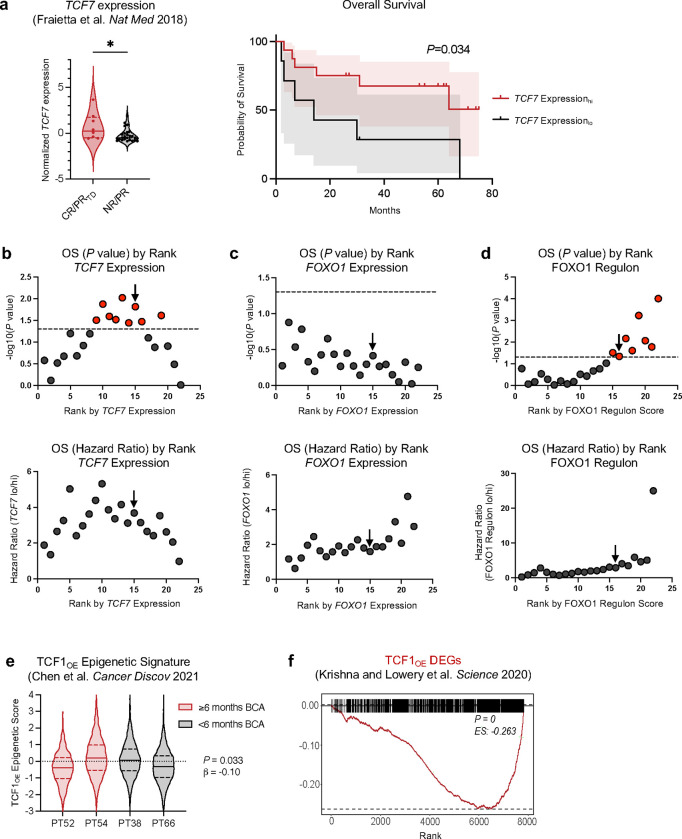
Extended Data Figure 8: Endogenous TCF7 transcript and FOXO1 regulon, but not TCFOE transcriptional or epigenetic signatures, predict CAR T and TIL responses in patients.
a, Single-sample gene set enrichment analyses (GSEA) were performed on RNA-sequencing data generated from ex vivo CAR-stimulated patient CTL019 T cells published in Fraietta et al. 2018 (cite). Enrichment score stratification points for patient survival analyses were determined using previously published methods (Jung et al 2023). TCF7 transcript correlates with response to CAR T (left) and overall survival (right). b-d, P values (top) and hazard ratios (bottom) of different stratification points in relation to overall survival (OS) of TCF7 expression (b), FOXO1 expression (c), and FOXO1 regulon (d). Dotted lines are drawn at P < 0.05, and black arrows indicate the stratification points used. e, Epigenetic signatures derived from differentially accessible peaks (P < 0.05) in CD19.28ζ TCF1OE CD8+ cells vs. tNGFR controls (Extended Data Fig. 5) were applied to scATAC-seq data generated from B-ALL apheresed patient T cells published in Chen et al6. The TCF1OE-derived epigenetic signature was not associated with patients with durable CAR T persistence (≥ 6 months B cell aplasia, BCA; Patient 52, n = 616 cells; Patient 54, n = 2959 cells) compared to those with short persistence (< 6 months BCA, Patient 38, n = 2093 cells; Patient 66, n = 2355 cells). f, GSEA analyses were performed on DEGs from CD39−CD69− patient TIL that correlated with responses in adult melanoma (Krishna and Lowery et al). HA.28ζ TCF1OE CD8+ DEGs (Fig. 3) were significantly de-enriched in CD39−CD69− TIL. a, Mann-Whitney test (left), Mantel-Cox test (right); e, Wald test of a linear regression model.