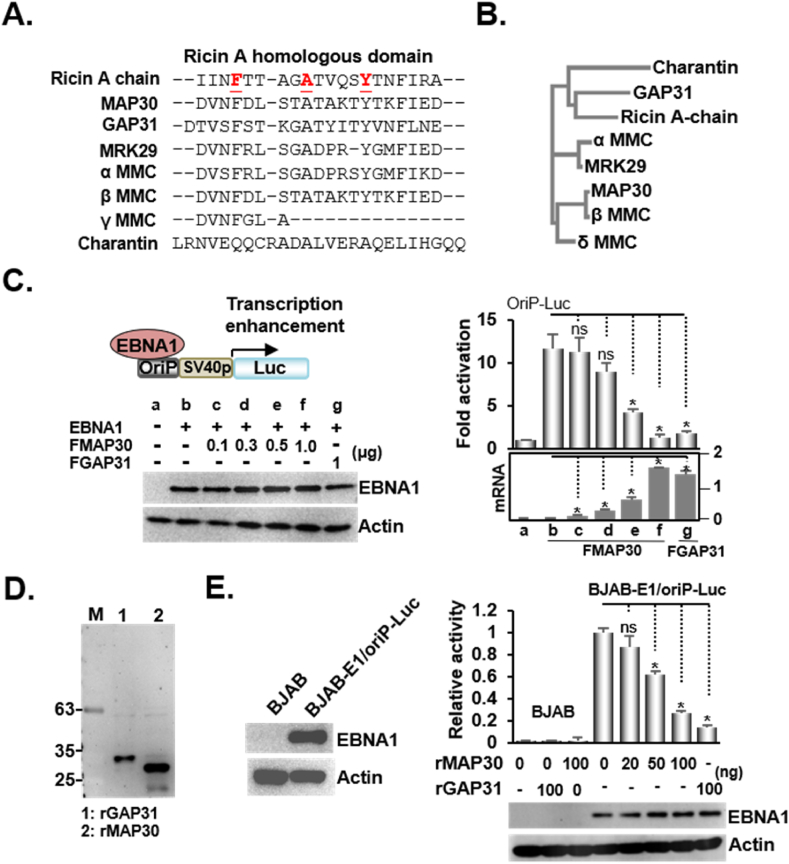
Fig. 1.
MAP30 inhibits EBNA1/oriP-Luc mediated transcription enhancement A). This figure shows the N-terminal alignment of the ricin-like proteins and the ricin N-terminal peptide encompassing amino residues from 8 to 22 was used as the template. The conserved residues of phenylalanine, alanine, and tyrosine are underlined and shown in boldface marked with red color. B). The phylogenetic tree depicts the lines of evolutionary descent of N-termini from different ricin-like proteins. C). A transfection-mediated EBNA1/oriP-Luc induced luciferase reporter assay was performed using the Flag-tagged DYKDDDDK expression plasmids of EBNA1(FEBNA1), MAP30(FMAP30), and GAP31(FGAP31), and the reporter plasmids, oriP-Luc, and β-Gal internal control. The resulting fold activation in each sample was determined by expressing the luciferase activity to β-Gal activity. The relative mRNA levels produced by 0.1–1 μg FMAP30 or 1 μg of GAP31 to GAPDH internal control and the immune blot images for FEBNA1 and actin control are also shown. D). The recombinant proteins rGAP31 and rMAP30 were purified and resolved by Western blot. A student t-test was performed to compare the resulting fold activation in each FMAP30 co-transfected sample to the EBNA1/oriP-Luc transfected sample. A p-value of <0.05 is considered statistically significant and an asterisk (*) is marked, whereas non-significance (ns) is noted when p > 0.05. The same approach was employed for all the necessary comparisons in all the following statistical analyses described in the study. E). Cells derived from an EBNA1/oriP-Luc reporter cell line (BJAB-E1/oriP-Luc) were treated with the indicated amount of rGAP31 or rMAP30 and subjected to a luciferase activity assay.