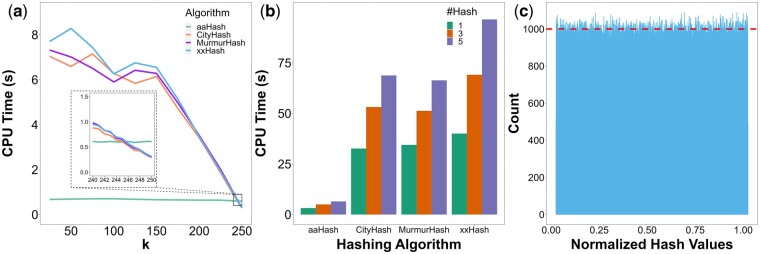
Figure 1.
Performance of aaHash. (a) Runtime for hashing 1 000 000 × 250 amino acids residue long sequences with k-mer lengths from 25 to 250. aaHash outperforms all other hashing methods when computing more than five subsequent k-mers (i.e. k < 246, see inset). (b) Comparing multi-hashing runtime of aaHash versus other state-of-the-art hashing functions for one billion 50-mers. aaHash hashing is ∼10× faster than the closest competitor, CityHash. The colours indicate the number of hashes generated. (c) Histogram of 1 000 000 100-mer hashes generated by aaHash from a random amino acid sequence of length 1 000 099. The dashed line indicates the average number of hashes in a bin (1000). The hash values were normalized by dividing the hash values by , the largest 64-bit integer, and plotted on the histogram with bin size of 1000. The mean and standard deviation of the bin counts are 1000.0 ± 31.4, demonstrating the empirical uniformity of aaHash.