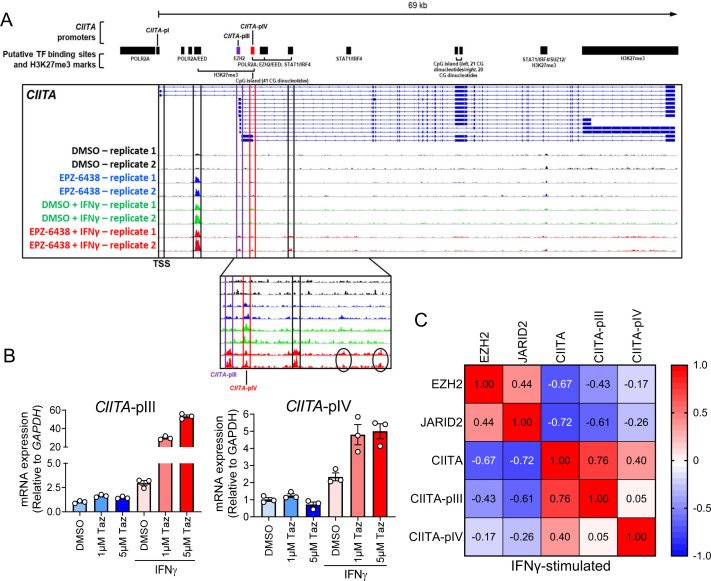
Figure 5.
Tazemetostat treatment exposes accessible chromatin at regions containing putative IRF1 and STAT1 binding motifs in IFN-γ-stimulated CHL-1 cells. (A) CHL-1 cells were pretreated for 4 days with tazemetostat before 24 hours stimulation with IFN-γ. Cells were harvested and processed for assays for transposase-accessible chromatin with sequencing. Black bars represent predicted target sites for H3K27me3 marks, CpG islands, and promoters driving transcription of the indicated genes, based on the publicly available data from multiple tissue types and conditions in the UCSC database. Putative transcription factor (TF) binding sites for IFN-γ signaling (STAT1 and IRF4), based on predictions made using the https://molotool.autosome.org/and a permissive p value setting of 0.0005. (B) mRNA expression assessed by qPCR using primers specific for the pIII and pIV 5’UTR. (C) Correlation matrix for qPCR data across seven cell lines (A375, SKMEL5, COLO829, SKMEL28, MEWO, SKMEL1, and CHL1) under interferon-stimulated conditions. CIITA, class II major histocompatibility complex transactivator II; DMSO, dimthyl sulfoxide; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; IFN, interferon; JARID2, Jumoni and AT-rich interaction domain-containing; mRNA, messenger RNA; pIV, promoter IV; qPCR, quantitative polymerase chain reaction; USCS, University of California, Santa Cruz.