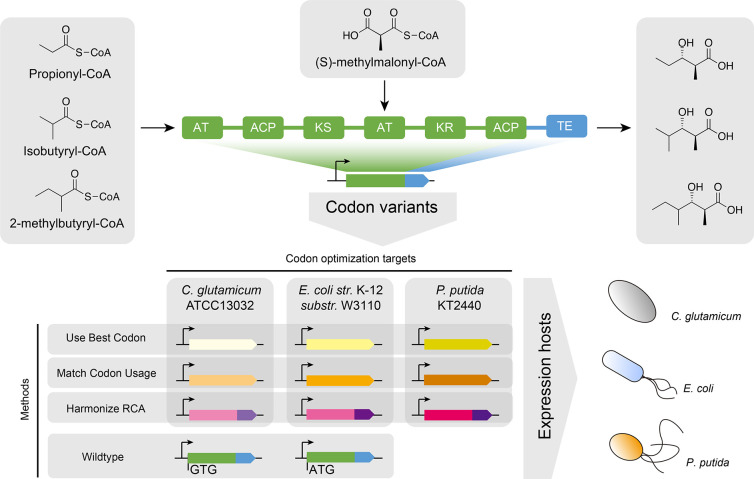
Figure 1.
Engineered polyketide synthase with applied codon optimization strategies and targeted heterologous hosts. The loading module and module 1 originates from the lipomycin polyketide synthase (LipPKS) from Streptomyces aureofaciens Tü117 (green). The thioesterase domain (TE) originates from the erythromycin PKS (EryPKS) from Saccharopolyspora erythraea NRRL2338 (blue). By fusion of these two parts together, the engineered PKS design yields a variety of short-chain 3-hydroxy acids. The gene sequence of the reprogrammed LipPKS was codon optimized using the DNA Chisel algorithms for “Use Best Codon” (yellow), “Match Codon Usage” (orange), and “Harmonize RCA” (red/purple). All algorithms preserve the amino acid sequence of the protein. The “Use Best Codon” method replaces each codon with the most frequently used codon. “Match Codon Usage” matches the codon frequency of the original codon sequence with the codon usage of the targeted host. “Harmonize RCA” applies and matches the codon frequency of the targeted host with the codon usage of the native host. The “Harmonize RCA” algorithm required codon optimization of the LipPKS and EryPKS parts separately. Codon optimizations targeted the three heterologous hosts C. glutamicum, E. coli, and P. putida.