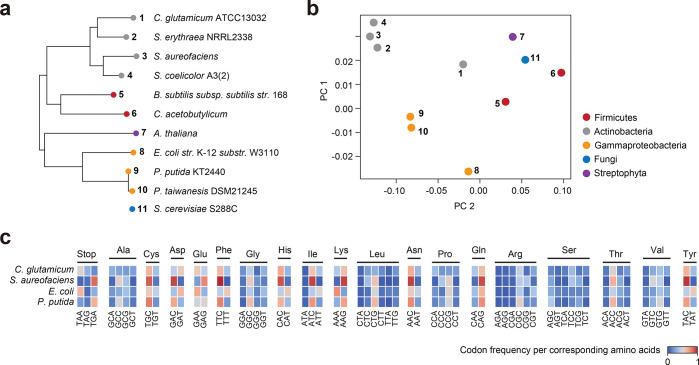
Figure 2.
Global analysis of codon usage preferences between different species. (a) 16S rRNA sequence-based phylogenetic analysis of targeted hosts, native polyketide synthase hosts, and outgroup references. Saccharomyces cerevisiae S288C was excluded. (b) Principal component analysis (PCA) of codon usage tables associated with the same species from the phylogenetic analysis. The first two principal components (PCs), accounting for the highest explained variance, were selected and visualized. (c) Comparison of the codon frequency per corresponding amino acid between C. glutamicum, S. aureofaciens, E. coli, and P. putida. The amino acids methionine and tryptophan are encoded by a singular codon and were excluded from this analysis.