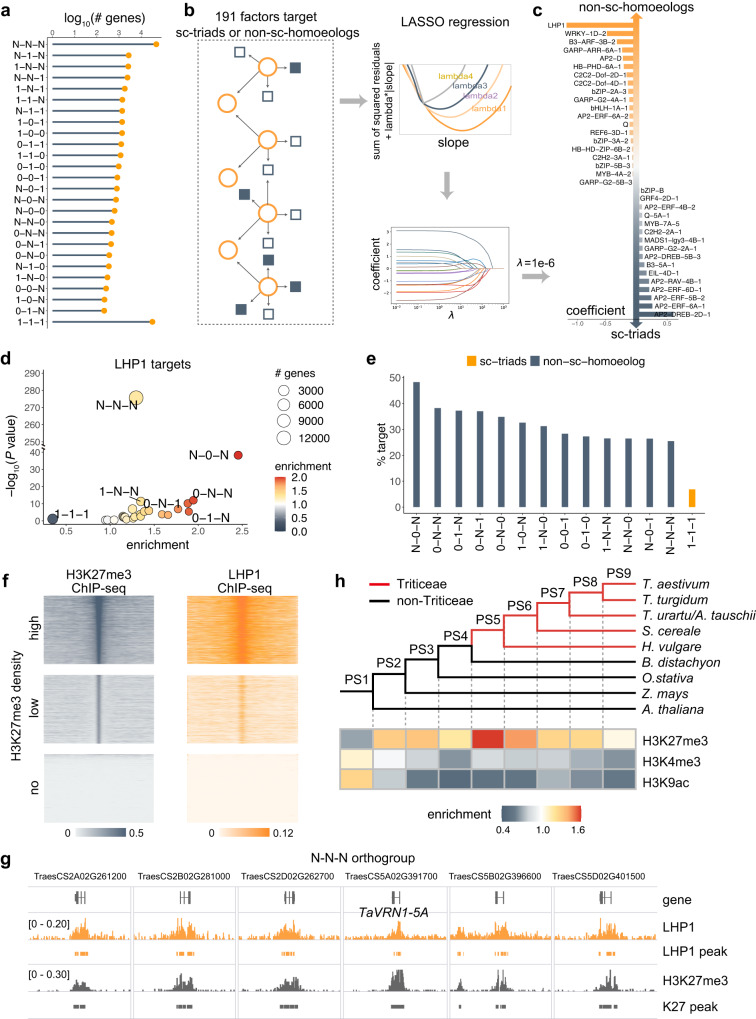
Fig. 1. Prediction of the core trans-factors regulating subgenome-diversified genes.
a Number of sc-triads (1:1:1) and non-sc-homoeologs with varying number of homologs across subgenomes in common wheat. b Workflow for predicting the core factors regulating sc-triads (1:1:1) and non-sc-homoeologs across subgenomes. c LASSO coefficients of the trans-factor binding-related regulation of non-sc-homoeologs (top) and sc-triad (bottom) genes. The coefficient value represents the relative importance of the trans-factor contributing to regulation. d Enrichment of different homoeologous groups among LHP1-targeted genes. e Fractions of homoeolog groups targeted by LHP1. f ChIP-seq read densities of LHP1 and H3K27me3 surrounding H3K27me3 peaks. g Genomic tracks illustrating the co-occupation of LHP1 and H3K27me3 ChIP-seq signals surrounding VRN1-5A and its homoeologs. h Enrichment of the typical epigenetic marks in eight phylostrata corresponding to the phylogenetic internodes, which are bordered by vertical grids and denote sets of genes whose founder genes originated in the corresponding evolutionary periods (e.g., the left-most phylostratum represents the common ancestor of dicots and monocots). The color represents the enrichment score. High enrichment scores reflect the expansion of the targets with a given epigenetic mark in the corresponding phylostrata.