Fig. 6. Identification of a PA binding pocket in Smo.
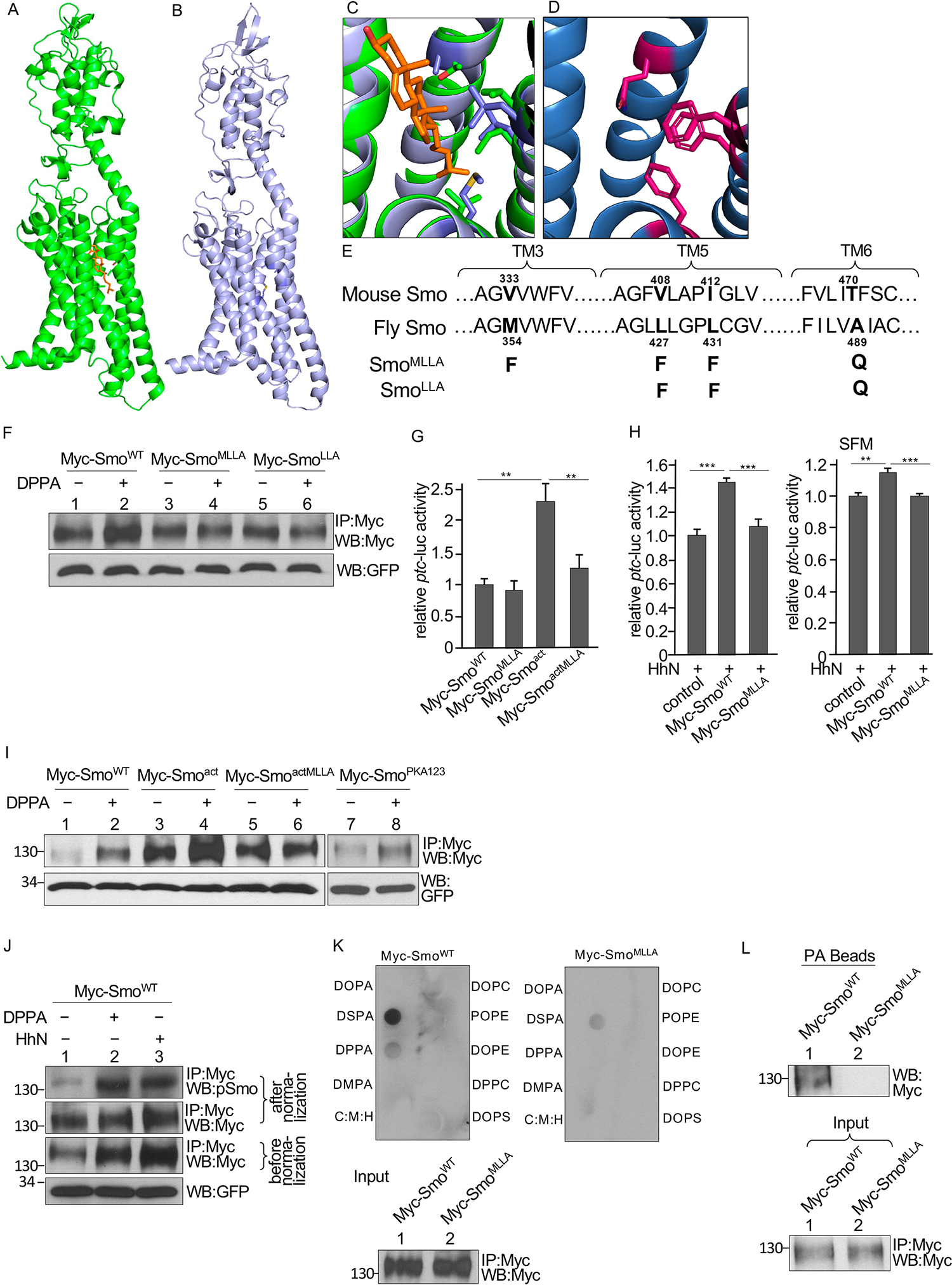
(A) Structure of mouse Smo with the cholesterol molecule in orange. (B) Alphafold model of Drosophila Smo. (C) The sterol binding site of mouse Smo (green) with the Drosophila model (blue) superimposed. (D) Alphafold model of the sterol binding site in the Drosophila SmoMLLA mutant. (E) Residues in Smo TM3, TM5, and TM6 domains that contribute to a pocket located near the sterol binding site and predicted to bind PA. Sequences of Drosophila and mouse Smo are shown. Mutations tested in fly Smo are indicated. (F) Myc immunoprecipitates (IP) from S2 cells transfected with either WT Smo or the indicated mutant forms of Smo and treated with DPPA were blotted (WB) for Myc and GFP. GFP served as transfection and lysate control. N=3 independent experiments. Statistical analysis of Smo abundance is shown in the Supplementary Materials (fig. S6B). (G) Quantification of ptc-luc reporter activity in S2 cells were cotransfected with the reporter, Ci, and the indicated Smo constructs. N=3 independent experiments. ** indicates a p < 0.01, versus control (Student’s t test). (H) Quantification of ptc-luc reporter activity in S2 cells cotransfected with the reporter, Ci, HhN, and the indicated Smo constructs and cultured in either regular medium or serum-free medium (SFM). N=3 independent experiments. *** indicates a p < 0.001, versus control; ** indicates a p < 0.01, versus control (Student’s t test). (I) Myc immunoprecipitates (IP) from S2 cells transfected with the indicated Smo constructs and treated with DPPA were blotted for Myc and GFP. N=3 independent experiments. Statistical analysis of Smo abundance is shown in the Supplementary Materials (fig. S6C). (J) Myc immunoprecipitates from S2 cells transfected with Myc-SmoWT and either cotransfected with HhN or treated with DPPA were blotted for Myc and phosphorylated Smo (pSmo). Samples were normalized by adjusting loading to quantify Smo phosphorylation fig. S6D). N=3 independent experiments. (K) Smo interaction with lipids in a solid phase lipid binding assay. Membranes dotted with the indicated lipids were incubated with Myc-SmoWT or Myc-SmoMLLA immunoprecipitated from S2 cells and immunoblotted for Myc. C:M:H is a vehicle control. The amounts of Myc-SmoWT and Myc-SmoMLLA added to each membrane (Input) are shown in the lower blot. N=3 independent experiments. (L) Myc immunoprecipitates of lysates from S2 cells expressing Myc-SmoWT or Myc-SmoMLLA were incubated with PA beads followed by Western blotting for Myc antibody to detect the bound Smo. The amounts of each immunoprecipitated protein applied to the beads (Input) is shown in the lower blot. N=3 independent experiments.
