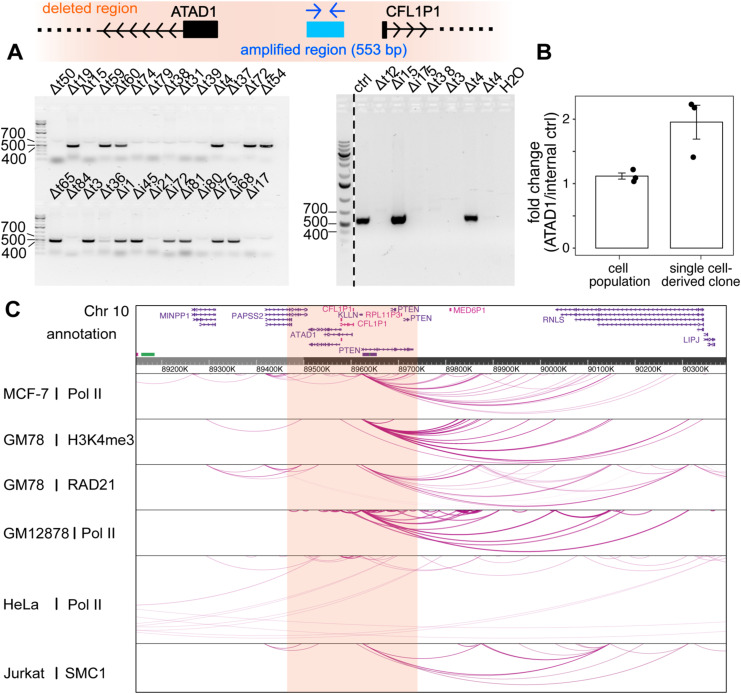
Figure S2. The loss of PAPSS2-PTEN locus verified in CRISPR-Cas9–modified single-cell–derived HAP1 clones and in the unmodified HAP1 cell population potentially affects interactions with nearby genomic regions.
(A) Schematic illustration shows the annealed region of PCR primers and amplicon length designed to validate the presence (553 bp PCR product) or absence (no PCR product) of the PAPSS2-PTEN locus (top) in HAP1 Δt and Δi cell clones. Agarose gel confirms the size of the obtained PCR products (bottom). (B) The barplot illustrates the relative frequencies of the PAPSS2-PTEN locus in genomic DNA molecules extracted from HAP1 cells (left) and single-cell–derived clones (right) with the PAPSS2-PTEN locus. The frequencies are presented as fold changes, obtained by normalizing the number of molecules carrying a genomic region in the ATAD1 promoter region to the number of molecules carrying our internal control (n = 3, mean ± SD, P = 0.08). (C) The hg38 Genome Browser displays long-range interaction loops centered on the PAPSS2-PTEN locus from publicly available Pol II, RAD21, H3K4me3, and SMC1 ChIA-PET data in multiple human cell lines.