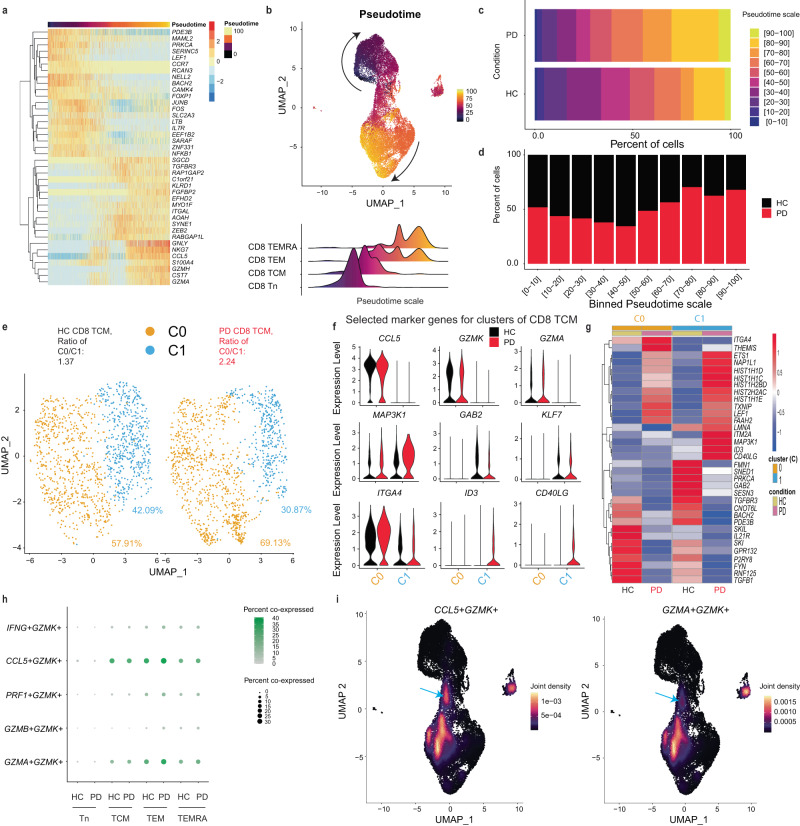
Fig. 7. scRNA-seq discloses the accelerated CD8 differentiation process in early-to-mid stage iPD.
a Heatmap showing the top 20 most-increased and -decreased genes along the predicted pseudotime trajectory ranked using log2FC (if p-value < 0.01) (n = ~25,000 cells). Each column represents one single cell. b Two-dimensional representation showing the CD8 memory T-cell differentiation trajectory using UMAP (upper). Lower, the ridge plots of pseudotime distribution among different CD8 subsets along differentiation trajectory. c Percentages of each binned psedutotime window along the pseudotime trajectory in iPD (n = ~12,000 cells) or HC (n = ~13,000 cells). d Stacked barplot showing the relative fraction of cells at the given predicted differentiation stage (during the given pseudotime window) between iPD and HC. e Unsupervised clustering analysis of CD8 TCM. Percentages of the two clusters and the ratios between two clusters were displayed. For comparability in visualization, cell numbers were randomly down-sampled to the same number for different conditions and subsets. f Violin plots of selected marker genes distinguishing different clusters within CD8 TCM. g Heatmap of selected top up- or downregulated genes in CD8 TCM from iPD vs HC. h Balloon plot showing the percentage of cells co-expressing the indicated markers in the given subset. If a cell shows the read count equal to or higher than 1 for each of the markers in the indicated combination, it is regarded as the cell co-expressing the set of markers. i UMAP plot showing joint density of CCL5 and GZMK (left) and of GZMA and GZMK (right) in all the individual CD8 T cells. Arrows refer to the area of TCM co-expressing the indicated markers. For a, the one-tailed ANOVA method was used to test whether any of the spline coefficients (for pseudotime fitting) is non-zero. In g, differential expression was analyzed using two-tailed nonparametric Wilcoxon Rank Sum test based on Bonferroni correction (only those with adjusted P-value ≤0.05 were considered). Each dot represents one single cell in (b, e, i). FC fold change, HC healthy controls, PD patients with Parkinson’s disease, UMAP uniform manifold approximation and projection.